Class 0: Introduction to the Course, Github, Computing Setup#
Goal of today’s class:
Introduce the syllabus
Ensure we’re all on the Discovery Cluster
Set up classroom virtual environment
Welcome to PHYS 7332 - Network Science Data II
How The Course Works#
This course offers an introduction to network analysis and is designed to provide students with an overview of the core data scientific skills required to analyze complex networks. Through hands-on lectures, labs, and projects, students will learn actionable skills about network analysis techniques using Python (in particular, the networkx library). The course network data collection, data input/output, network statistics, dynamics, and visualization. Students also learn about random graph models and algorithms for computing network properties like path lengths, clustering, degree distributions, and community structure. In addition, students will develop web scraping skills and will be introduced to the vast landscape of software tools for analyzing complex networks. The course ends with a large-scale final project that demonstrates the proficiency of the students in network analysis.
This syllabus may be updated and can be found here: https://brennanklein.com/phys7332-fall24.
This course has been built from the foundation of the years of work/development by Matteo Chinazzi and Qian Zhang, for earlier iterations of Network Science Data.
Course Learning Outcomes#
Proficiency in Python and
networkxfor network analysis.Strong foundation of complex network algorithms and their applications.
Skills in statistical description of networks.
Experience in collecting and analyzing online data.
Broad knowledge of various network libraries and tools.
Course Materials#
There are no required materials for this course, but we will periodically draw from:
Bagrow & Ahn (2024). Working with Network Data: A Data Science Perspective. Cambridge University Press; 1st Edition; 978-1009212595. https://www.cambridge.org/network-data.
Additionally, we recommend engagement with other useful network science and/or Python materials:
Barabási (2016). Network Science. Cambridge University Press; 1st Edition; 978-1107076266. http://networksciencebook.com/.
Newman (2018). Networks: An Introduction. Oxford University Press; 2nd Edition; 978-0198805090. https://global.oup.com/academic/product/networks-9780198805090.
Barrat, Barthelemy & Vespignani (2008). Dynamical Processes on Complex Networks. Cambridge University Press; 1st Edition; 978-0511791383. https://doi.org/10.1017/CBO9780511791383.
VanderPlas (2019). Python Data Science Handbook. O’Reilly Media, Inc; 978-1491912058. jakevdp/PythonDataScienceHandbook.
Class Expectations#
This is a “new” class (it had previously been taught as a part of the Network Science PhD program at Northeastern, but for the past 5+ years we’ve not taught it). We’ve brought it back after years of realizing that it contains valuable skills for students studying network science, and we’re hoping to gain important insights around what skills are essential to teach, what can be assumed prior knowledge, and what is too challenging for students. As such, we’ll be asking for frequent feedback on the assignments, classes, etc.
Classroom Interactions#
This is largely a “coding” course. That means we’ll all have our computers out and follow along the same notebook as the instructor is teaching. There will be frequent “your turn” sections of the code, where we’ll expect you to break off into solo / small teams to solve an in-class challenge. When the class comes back together, we’ll ask for volunteers to share screen and show off what they’ve done.
Coursework, Class Structure, Grading#
This is a twice-weekly hands-on class that emphasizes building experience with coding. This does not necessarily mean every second of every class will be live-coding, but it will inevitably come up in how the class is taught. We are often on the lookout for improving the pedagogical approach to this material, and we would welcome feedback on class structure. The course will be co-taught, featuring lectures from the core instructors as well as outside experts. Grading in this course will be as follows:
Class Attendance & Participation: 10%
Problem Sets: 45%
Mid-Semester Project Presentation: 15%
Final Project — Presentation & Report: 30%
Final Project#
The final project for this course is a chance for students to synthesize their knowledge of network analysis into pedagogical materials around a topic of their choosing. Modeled after chapters in the Jupyter book for this course, students will be required to make a new “chapter” for our class’s textbook; this requires creating a thoroughly documented, informative Python notebook that explains an advanced topic that was not deeply explored in the course. For these projects, students are required to conduct their own research into the background of the technique, the original paper(s) introducing the topic, and how/if it is currently used in today’s network analysis literature. Students will demonstrate that they have mastered this technique by using informative data for illustrating the usefulness of the topic they’ve chosen. Every chapter should contain informative data visualizations that build on one another, section-by-section. The purpose of this assignment is to demonstrate the coding skills gained in this course, doing so by learning a new network analysis technique and sharing it with members of the class. Over time, these lessons may find their way into the curriculum for future iterations of this class. Halfway through the semester, there will be project update presentations where students receive class and instructor feedback on their project topics. Throughout, we will be available to brainstorm students’ ideas for project topics.
Ideas for Final Project Chapters (non-exhaustive):#
Graph Embedding (or other ML technique)
Network Reconstruction from Dynamics
Link Prediction
Graph Distances and Network Comparison
Motifs in Networks
Network Sparsification
Spectral Properties of Networks
Mechanistic vs Statistical Network Models
Robustness / Resilience of Network Structure
Network Game Theory (Prisoner’s Dilemma, Schelling Model, etc.)
Homophily in Networks
Network Geometry and Random Hyperbolic Graphs
Information Theory in/of Complex Networks
Discrete Models of Network Dynamics (Voter model, Ising model, SIS, etc.)
Continuous Models of Network Dynamics (Kuramoto model, Lotka-Volterra model, etc.)
Percolation in Networks
Signed Networks
Coarse Graining Networks
Mesoscale Structure in Networks (e.g. core-periphery)
Graph Isomorphism and Approximate Isomorphism
Inference in Networks: Beyond Community Detection
Activity-Driven Network Models
Forecasting with Networks
Higher-Order Networks
Introduction to Graph Neural Networks
Hopfield Networks and Boltzmann Machines
Graph Curvature or Topology
Reservoir Computing
Adaptive Networks
Multiplex/Multilayer Networks
Simple vs. Complex Contagion
Graph Summarization Techniques
Network Anomalies
Modeling Cascading Failures
Topological Data Analysis in Networks
Self-organized Criticality in Networks
Network Rewiring Dynamics
Fitting Distributions to Network Data
Hierarchical Networks
Ranking in Networks
Deeper Dive: Random Walks on Networks
Deeper Dive: Directed Networks
Deeper Dive: Network Communities
Deeper Dive: Network Null Models
Deeper Dive: Network Paths and their Statistics
Deeper Dive: Network Growth Models
Deeper Dive: Network Sampling
Deeper Dive: Spatially-Embedded and Urban Networks
Deeper Dive: Hypothesis Testing in Social Networks
Deeper Dive: Working with Massive Data
Deeper Dive: Bipartite Networks
Many more possible ideas! Send us whatever you come up with
What You’ll Learn#
Students should leave this class with an ever-growing codebase of resources for analyzing and deriving insights from complex networks, using Python. These skills range from being able to (from scratch) code algorithms on graphs, including path length calculations, network sampling, dynamical processes, and network null models; as well as interfacing with standard data science questions around storing, querying, and analyzing large complex datasets.
Schedule#
DATE |
CLASS |
|---|---|
Wed, Sep 4, 24 |
Class 0: Introduction to the Course, Github, Computing Setup |
Thu, Sep 5, 24 |
Class 1: Python Refresher (Data Structures, Numpy) |
Fri, Sep 6, 24 |
— |
Wed, Sep 11, 24 |
Class 2: Introduction to Networkx 1 — Loading Data, Basic Statistics |
Thu, Sep 12, 24 |
Class 3: Introduction to Networkx 2 — Graph Algorithms |
Fri, Sep 13, 24 |
Announce Assignment 1 |
Wed, Sep 18, 24 |
Class 4: Distributions of Network Properties & Centralities |
Thu, Sep 19, 24 |
Class 5: Scraping Web Data 1 — BeautifulSoup, HTML, Pandas |
Fri, Sep 20, 24 |
— |
Wed, Sep 25, 24 |
Class 6: Scraping Web Data 2 — Creating a Network from Scraped Data |
Thu, Sep 26, 24 |
Class 7: Big Data 1 — Algorithmic Complexity & Computing Paths |
Fri, Sep 27, 24 |
Assignment 1 due September 27 |
Wed, Oct 2, 24 |
Class 8: Data Science 1 — Pandas, SQL, Regressions |
Thu, Oct 3, 24 |
Class 9: Data Science 2 — Querying SQL Tables for Network Construction |
Fri, Oct 4, 24 |
Announce Assignment 2 |
Wed, Oct 9, 24 |
Class 10: Clustering & Community Detection 1 — Traditional |
Thu, Oct 10, 24 |
Class 11: Clustering & Community Detection 2 — Contemporary |
Fri, Oct 11, 24 |
— |
Wed, Oct 16, 24 |
Class 12: Visualization 1 — Python + Gephi |
Thu, Oct 17, 24 |
Class 13: Project Update Presentations |
Fri, Oct 18, 24 |
Assignment 2 due October 18 |
Wed, Oct 23, 24 |
Class 14: Introduction to Machine Learning 1 — General |
Thu, Oct 24, 24 |
Class 15: Introduction to Machine Learning 2 — Networks |
Fri, Oct 25, 24 |
Announce Assignment 3 |
Wed, Oct 30, 24 |
Class 16: Visualization 2 — Guest Lecture (Pedro Cruz, Northeastern) |
Thu, Oct 31, 24 |
Class 17: Dynamics on Networks 1 — Diffusion and Random Walks |
Fri, Nov 1, 24 |
— |
Wed, Nov 6, 24 |
Class 18: Dynamics on Networks 2 — Compartmental Models |
Thu, Nov 7, 24 |
Class 19: Dynamics on Networks 3 — Agent-Based Models |
Fri, Nov 8, 24 |
Assignment 3 due November 8 |
Wed, Nov 13, 24 |
Class 20: Big Data 2 — Scalability |
Thu, Nov 14, 24 |
Class 21: Network Sampling |
Fri, Nov 15, 24 |
— |
Wed, Nov 20, 24 |
Class 22: Network Filtering/Thresholding |
Thu, Nov 21, 24 |
Class 23: Dynamic of Networks: Temporal Networks |
Fri, Nov 22, 24 |
— |
Wed, Nov 27, 24 |
Thanksgiving Break (No Class) |
Thu, Nov 28, 24 |
— |
Fri, Nov 29, 24 |
— |
Wed, Dec 4, 24 |
Class 24: Instructor’s Choice: Spatial Data, OSMNX, GeoPandas |
Thu, Dec 5, 24 |
Class 25: Wiggle Room Class / Office Hours |
Fri, Dec 6, 24 |
— |
Wed, Dec 11, 24 |
Class 26: Final Presentations |
Discovery Cluster#
From Research Computing at Northeastern: “The Northeastern University HPC Cluster is a high-powered, multi-node, parallel computing system designed to meet the computational and data-intensive needs of various academic and industry-oriented research projects. The cluster incorporates cutting-edge computing technologies and robust storage solutions, providing an efficient and scalable environment for large-scale simulations, complex calculations, artificial intelligence, machine learning, big data analytics, bioinformatics, and more.
Whether you are a seasoned user or just beginning your journey into high-performance computing, our portal offers comprehensive resources, including in-depth guides, tutorials, best practices, and troubleshooting tips. Furthermore, the platform provides a streamlined interface to monitor, submit, and manage your computational jobs on the HPC cluster.
We invite you to explore the resources, tools, and services available through the portal. Join us as we endeavor to harness the power of high-performance computing, enabling breakthroughs in research and fostering innovation at Northeastern University.” (https://rc-docs.northeastern.edu/en/latest/index.html)
Documentation:#
Getting started: https://rc-docs.northeastern.edu/en/latest/gettingstarted/index.html
Trainings:#
Video and tutorial files for getting started on the Discovery cluster: https://rc.northeastern.edu/support/training/
Open OnDemand (OOD)#
“Open OnDemand empowers students, researchers, and industry professionals with remote web access to supercomputers.” (https://openondemand.org/)
Open OnDemand is a software platform that provides an easy-to-use web interface for accessing and managing high-performance computing (HPC) resources. It simplifies the process of submitting jobs, managing files, and running interactive applications on HPC systems without the need for complex command-line interactions. For the class this fall, we’ll be using Open OnDemand to give students hands-on experience with HPC resources, letting us run simulations, analyze large datasets, and explore computational tools in a user-friendly environment. This is designed to let students focus on learning and applying computational techniques without getting bogged down by technical complexities.
Interacting with OOD#
Open OnDemand Documentation: https://osc.github.io/ood-documentation/latest/
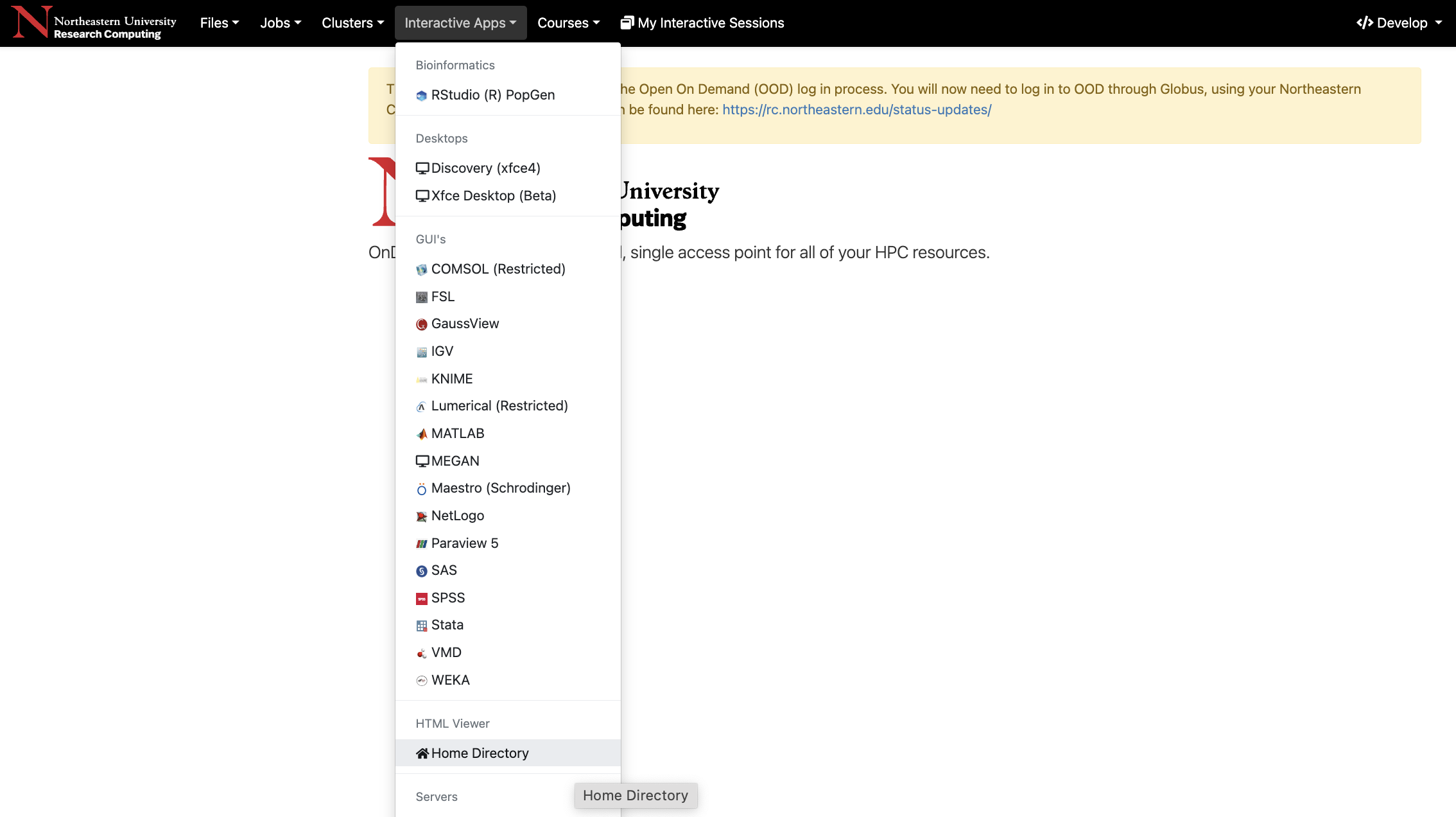
Interactive Apps:
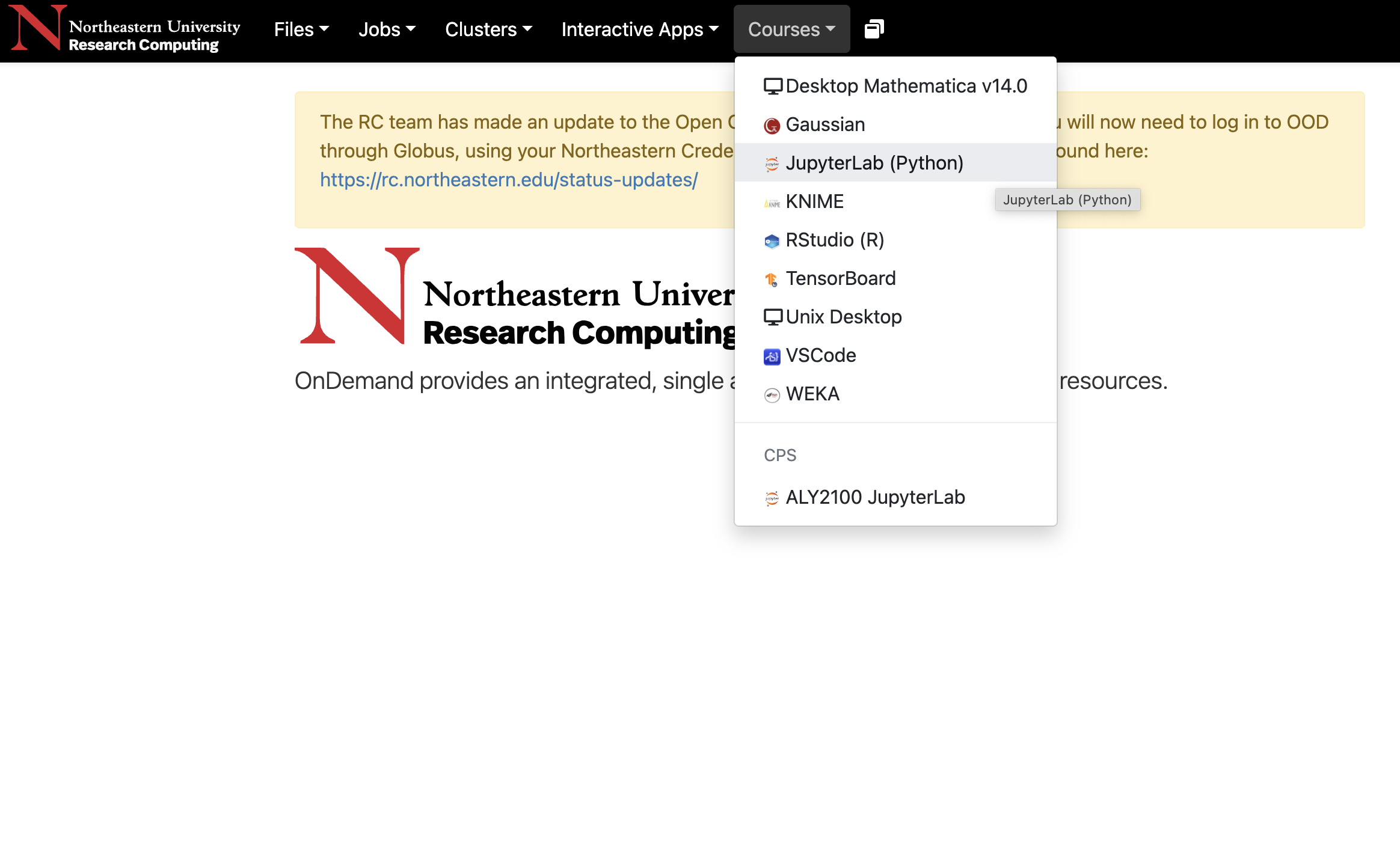
Courses:
Our Very Own Partition#
Jupyter Notebook Server#
This app will launch JupyterLab Notebook on a node on the courses partition, with requested CPUs, GPUs and memory for up to 24 hours. A custom conda environment is optional.
Virtual Environment#
What is conda and why do we use it?#
Python has the ability to do a lot of things on its own, but sometimes we need to install packages that extend Python’s functionality. For example, scikit-learn has a lot of machine learning algorithms, and geopy does handy geographic math (like calculating geodesic distances). Managing the dependencies these packages have on other packages is really annoying and really easy to mess up (in fact, it is NP-complete,, which means it’s a very hard type of computational problem), so we use tools called package managers. pip is one package manager, and conda is a fancier one. conda, short for anaconda, can handle packages that are outside the Python ecosystem, like C compilers, whereas pip cannot.
Class Virtual Environment#
Our class has a shared virtual environment that you can access once you use module to load conda. module is a tool that’s frequently used on computing clusters to load software packages on demand. Here’s some information on loading conda on Discovery, and here’s some information on module. First, you need to request a compute node on Discovery. We’ll go to Open On Demand (https://ood.discovery.neu.edu) and open up an interactive shell:
Once you’re in the interactive shell, request a compute node:
srun --partition=short --nodes=1 --cpus-per-task=1 --pty /bin/bash
And then you’ll load conda:
module load anaconda3/2022.05
From here, we can load the class virtual environment:
source activate /courses/PHYS7332.202510/shared/phys7332-env
You should see (/courses/PHYS7332.202510/shared/phys7332-env) in front of your command prompt in the Discovery terminal.
GitHub#
What is GitHub?#
GitHub is a website that implements git. git is a piece of open-source software that does distributed version control. This is really important for big teams making changes simultaneously to a large codebase. Git lets people document and track their code, and its various versions, so they can collaborate more efficiently.
On Github, you’ll keep your code in what’s called a repository. That’s a chunk of code that all works together. You can download a repository to your own computer, make changes to it, and push them to the repository on GitHub. Then your changes will be reflected in the version of the repository that’s on GitHub, and anyone can download the version with your changes in it.
Our class has a repository that’s available here. You will download a fork of the repository to make your very own. Occasionally we’ll update the repository and you’ll need to incorporate the updates; we’ll explain how to do that in a second.
Forking and Maintaining The Class Repository#
First, let’s fork the repository. Go to the repository GitHub link. You’ll see a little icon on the top left that says “Fork”. Click on it:
Once you’ve clicked the fork button, you’ll get this screen:
 Make sure you’re listed as the owner and create the fork.
Then you’ll clone your fork in the Discovery terminal using the URL obtained via the green
Make sure you’re listed as the owner and create the fork.
Then you’ll clone your fork in the Discovery terminal using the URL obtained via the green Code buttion:
We’re going to first navigate to your folder in our course partition:
cd /courses/PHYS7332.202510/students/USERNAME where USERNAME is your Northeastern username. Then we’ll clone the repo:
git clone [URL YOU COPIED]
Now let’s set up a way to get updates from the original repo. Let’s cd into the repo:
cd REPO_FILEPATH
git remote add upstream https://github.com/asmithh/student-network-science-data-book.git
git fetch upstream
git checkout main
git merge upstream/main
This sets up the original version of the GitHub repo as an upstream source. Now we can, at any point, fetch the upstream source and merge it into our main branch, which is the main branch of your forked repository. You’ll be working in this forked repository in your /courses/PHYS7332.202510/students/USERNAME directory throughout the course. If you want to sync your forked version with the original version (as we will be adding new notebooks over the course of the class), you can run the following commands:
git fetch upstream
git checkout main
git merge upstream/main
(Here’s a StackOverflow thread explaining this in more detail.)
Final step: Start a Jupyter Lab instance!#
Recall from earlier, the “Courses” tab:

Click on that, then fill in the following information (after you do this once, it’ll auto-populate for future classes):
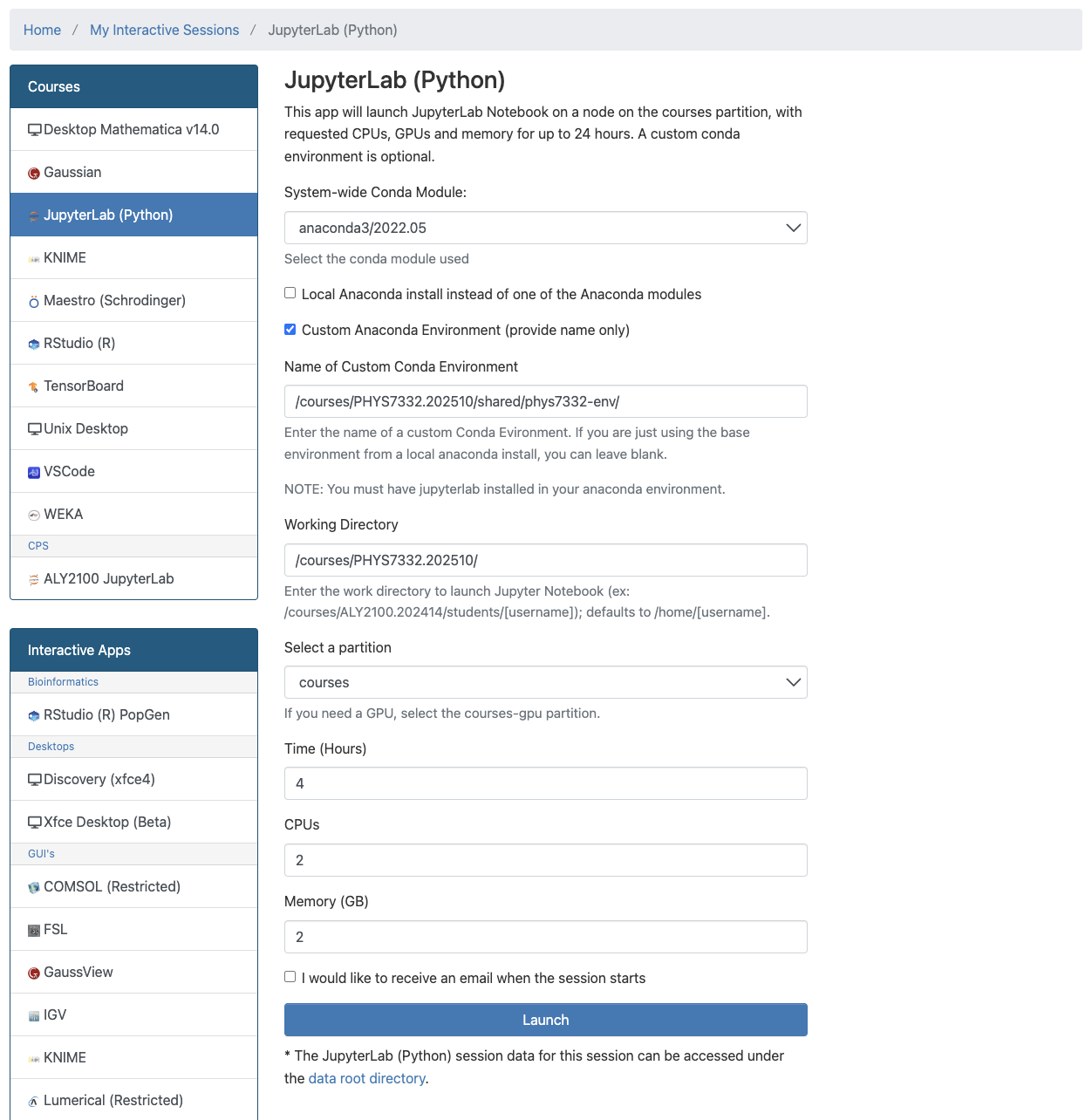
We’re in!
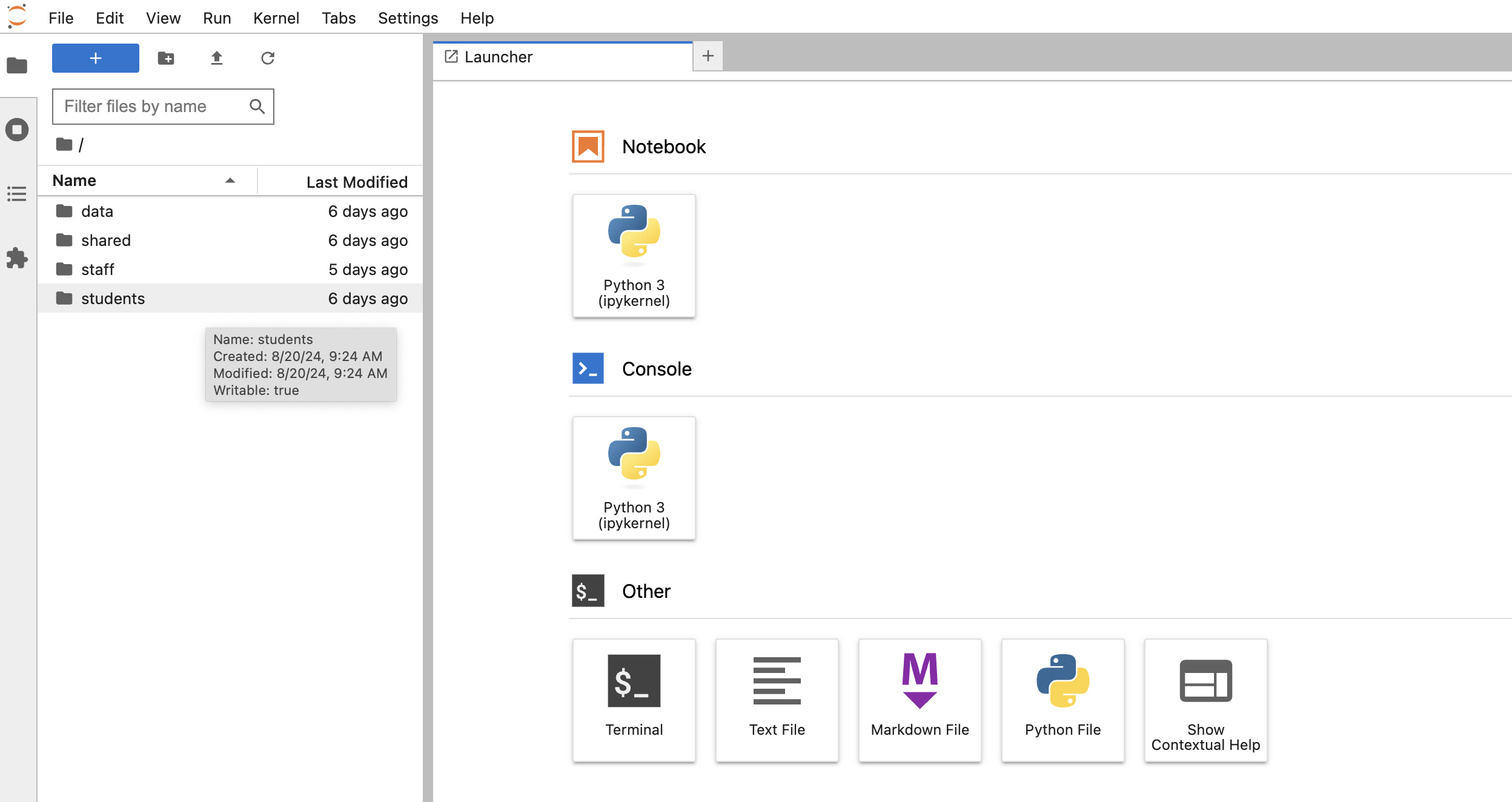
Now, navigate to this jupyter notebook, and get a feel for the notebooks, the folders, the files, etc. that you see.
Fixing Github Problems & Creating a One-Stop Shop For Git Updates#
We’re going to go through a series of steps to hopefully make updating the Github repo less of a pain in the butt in the long run!
Setting up SSH Keys with GitHub#
First we’re going to set up SSH keys on Discovery and put them on GitHub. This lets us authenticate our identity to GitHub and makes it possible for us to push our changed code to our forked repository. Neat! First, let’s follow the tutorial here to create SSH keys, because they explain it way better than I can.
Then you’ll add your public SSH key (the file that ends in .pub) to GitHub using these instructions. Label them something memorable, like discovery key, so you’ll know which machine(s) it’s attached to.
Syncing the Fork#
Now we’re going to sync our forked repository with the main student repository that I own. We can do this on GitHub, which is neat! This will incorporate all the fancy changes I made (okay, it’s basically one script) and let us test out the git update script. Go to your forked repository on GitHub; you’ll see a button that says Sync fork. Click on that; your screen should look something like this.
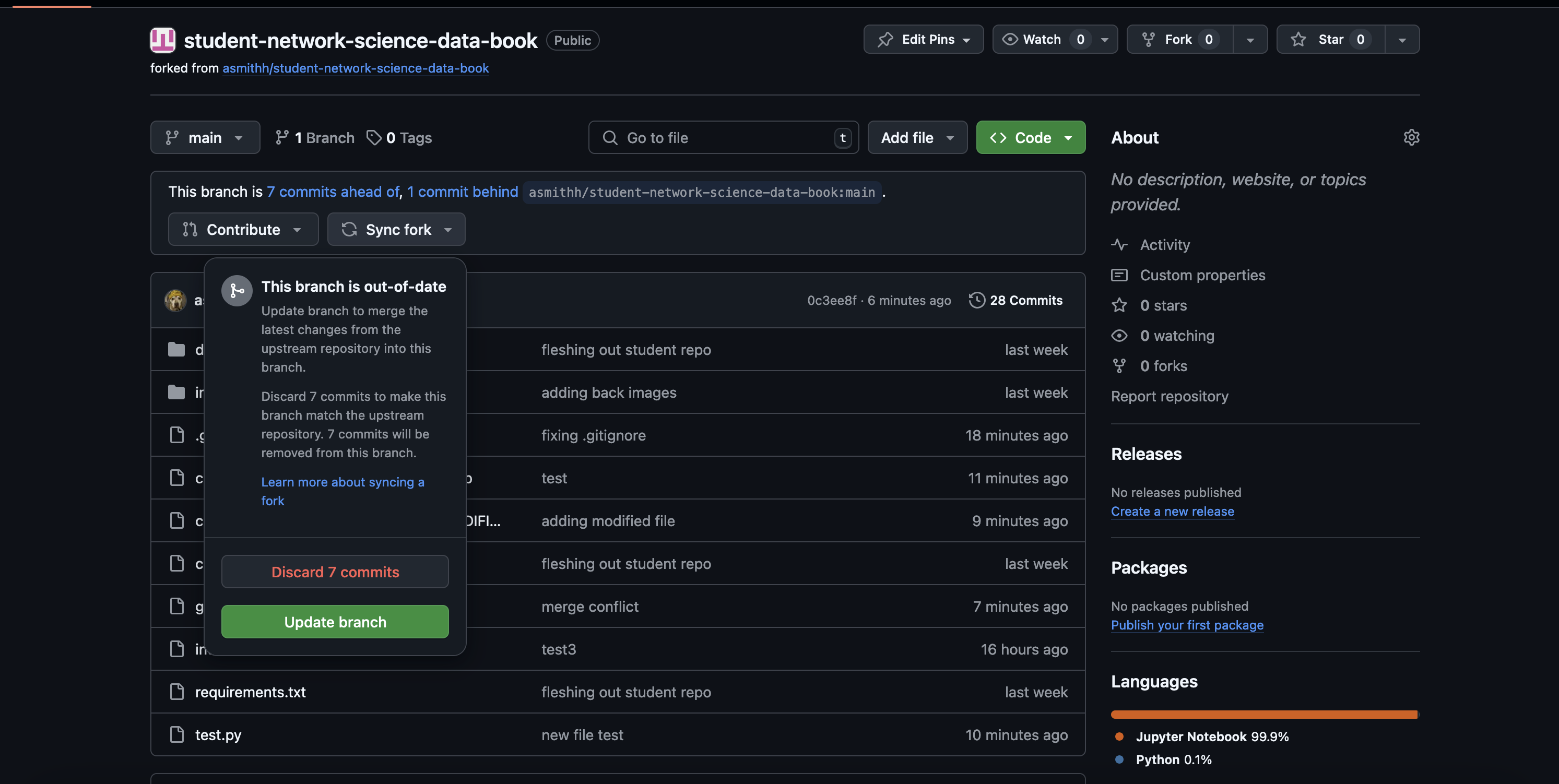
Click the green button (Update branch) to sync your fork with the main repository. Don’t discard any commits you may have made.
Going to Discovery and Trying This Weird Script#
Next we’ll go to Discovery and pull our new changes using git pull origin main. You should see a new file come up that is labeled git_fixer.py. That is going to (hopefully) be our new best friend for the near future.
Now, make some modifications to class_00_github_computing_setup.ipynb. You can just add comments at the end or get fancier; do what your heart desires.
Next, we’ll run python3 git_fixer.py. If this script works well enough, it should create a version of any files that you’ve modified from the original that have MODIFIED_{a bunch of numbers}.ipynb at the end of the filename. It will also update any files that have been modified on my end and add any new files that I’ve created in my repository.
Finally, run git status. You may need to add and commit the changes to your fork so you have your modified notebooks (ideally with your notes and added code) on GitHub in your forked repository. Then push them (git push origin main) so they’re reflected in your forked repository.
Going Forward#
At the beginning of each class, run git_fixer.py to make sure you have all the new code for the upcoming classes. Then you will run git status and add/commit/push any changes you wish to see reflected in your forked repository.
Next time…#
Python refresher! class_01_python_refresher.ipynb
References and further resources:#
Class Webpages
Jupyter Book: https://asmithh.github.io/network-science-data-book/intro.html
Syllabus and course details: https://brennanklein.com/phys7332-fall24
Discovery Cluster “OOD”: https://ood.discovery.neu.edu/
Open OnDemand Documentation: https://osc.github.io/ood-documentation/latest/
Python for R users: https://rebeccabarter.com/blog/2023-09-11-from_r_to_python