Class 23: Dynamic of Networks: Temporal Networks#
Goal of today’s class:
Create temporal networks from data
Learn key properties of temporal networks like inter-event time distributions, burstiness, etc.
Introduce the activity-driven network model
Acknowledgement: Some of the material in this lesson is based on a previous course offered by Matteo Chinazzi and Qian Zhang.
Come in. Sit down. Open Teams.
Make sure your notebook from last class is saved.
Open up the Jupyter Lab server.
Open up the Jupyter Lab terminal.
Activate Conda:
module load anaconda3/2022.05Activate the shared virtual environment:
source activate /courses/PHYS7332.202510/shared/phys7332-env/Run
python3 git_fixer2.pyGithub:
git status (figure out what files have changed)
git add … (add the file that you changed, aka the
_MODIFIEDone(s))git commit -m “your changes”
git push origin main
import networkx as nx
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
from matplotlib import rc
rc('axes', axisbelow=True)
rc('axes', fc='w')
rc('figure', fc='w')
rc('savefig', fc='w')
To begin: a challenge.#
In the data folder, there is a file sp.tsv.
Use
pandasto load this dataset.Name the columns [‘timestamp’, ‘node_i’, ‘node_j’]
Plot the number of connections between pairs of nodes per unit time.
You have until 11:15am to create a figure.
pass
pass
Temporal Networks (Time-Varying Networks)#
To motivate, what does contact tracing actually look like in an epidemic?#
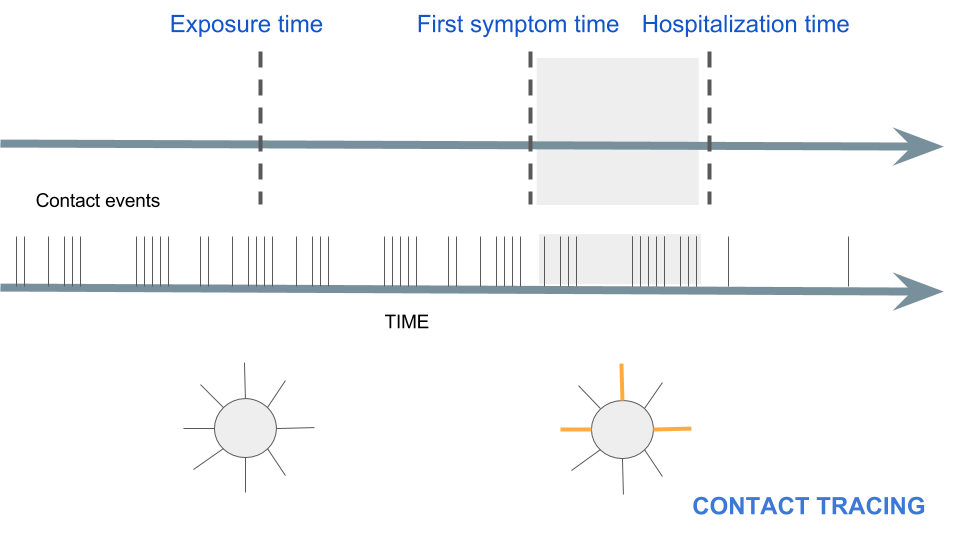
Temporal networks are everywhere#
Person-to-person communication
I. Emails
II.Mobile phone calls, or messages
III.Various online activities
One-to many communication
I. Twitter
II.Instagram
III.Wikipedia
Physical Proximity
I. Reality mining project
II. SocioPatterns project
Cell biology
I. Proteins interactions
II. Gene regulatory networks
III. Metabolic networks
Infrastructural Systems
I. Air transportation
II. Train routes
III. Bus routes
Neural and brain networks
I. Activation and correlation among different brain areas
II. Neuronal connectivity
Ecological systems
I. Food webs
II. Mobility and proximity of animals
Time is relative!#
\(t_G\) describes the evolution of the network
\(t_P\) describes the evolution of the process
scale of \(t_G/t_P\):
Representation#
Static network: adjacency matrix \(A_{ij}\)
Temporal network: adjacency matrix as a function of time \(A_{ij}(t)\)
Connected components#
I. Strongly connected: all \(i\) and \(j\) reachable within \(T\) (in both directions)
II. Weakly connected: all \(i\) and \(j\) reachable within \(T\) (in at least one direction)
Centralities#
I. Closeness: $\(c^{c}(i) = \frac{N-1}{\sum_{i\neq j} d_{ij}} \qquad \Rightarrow \qquad c^{c}(i, T) = \frac{N-1}{\sum_{i\neq j} \lambda_{ijT}}\)$
II. Betweenness:
\(g_{j,k}\) is the number of shortest paths between nodes \(j\) and \(k\)
\(g_{j,k}(i)\) is the number of shortest paths between nodes \(j\) and \(k\) going through \(i\)
let if \(j=k\), \(g_{jk} = 1\), and if \(i\in\{j,k\}\), \(g_{j,k}(i)=0\)
Loading temporal network data#
Revisiting the temporal network from above…
The High School Contact Network Dataset from the SocioPatterns project captures the temporal interactions between students in a French high school over the course of several days. The dataset was collected using wearable proximity sensors that recorded face-to-face interactions among students at 20-second intervals. Each interaction includes the IDs of the two individuals involved and the timestamp of the interaction in seconds since the start of the observation period.
This dataset is widely used in network science research to study temporal networks, social interactions, and spreading processes (e.g., information or disease propagation). Its temporal resolution and rich interaction data make it an excellent resource for exploring dynamic graph structures and understanding social behavior in structured environments like schools.
References
Cattuto, C., Van den Broeck, W., Barrat, A., Colizza, V., Pinton, J. F., & Vespignani, A. (2010). Dynamics of person-to-person interactions from distributed RFID sensor networks. PloS One, 5(7), e11596. https://doi.org/10.1371/journal.pone.0011596
Starnini, M., Baronchelli, A., & Pastor-Satorras, R. (2013). Modeling human dynamics of face-to-face interaction networks. Physical Review Letters, 110(16), 168701. https://doi.org/10.1103/PhysRevLett.110.168701
# time \t node_i \t node_j
tdf = pd.read_csv('data/sp.tsv',sep='\t',header=None,
names=['timestamp','node_i','node_j'])[['node_i','node_j','timestamp']]
bin_size = 60*1 # 1 minute
tdf["time_bin"] = (tdf['timestamp'] // bin_size) * bin_size
tdf
| node_i | node_j | timestamp | time_bin | |
|---|---|---|---|---|
| 0 | 1170 | 1644 | 0 | 0 |
| 1 | 1170 | 1613 | 20 | 0 |
| 2 | 1170 | 1644 | 260 | 240 |
| 3 | 1181 | 1651 | 380 | 360 |
| 4 | 1108 | 1190 | 460 | 420 |
| ... | ... | ... | ... | ... |
| 45042 | 880 | 887 | 729200 | 729180 |
| 45043 | 854 | 869 | 729200 | 729180 |
| 45044 | 880 | 887 | 729220 | 729180 |
| 45045 | 620 | 669 | 729380 | 729360 |
| 45046 | 669 | 677 | 729500 | 729480 |
45047 rows × 4 columns
aggregate_counts = tdf["time_bin"].value_counts().sort_index()
aggregate_counts
0 2
240 1
360 1
420 1
600 2
..
729000 4
729060 3
729180 7
729360 1
729480 1
Name: time_bin, Length: 4135, dtype: int64
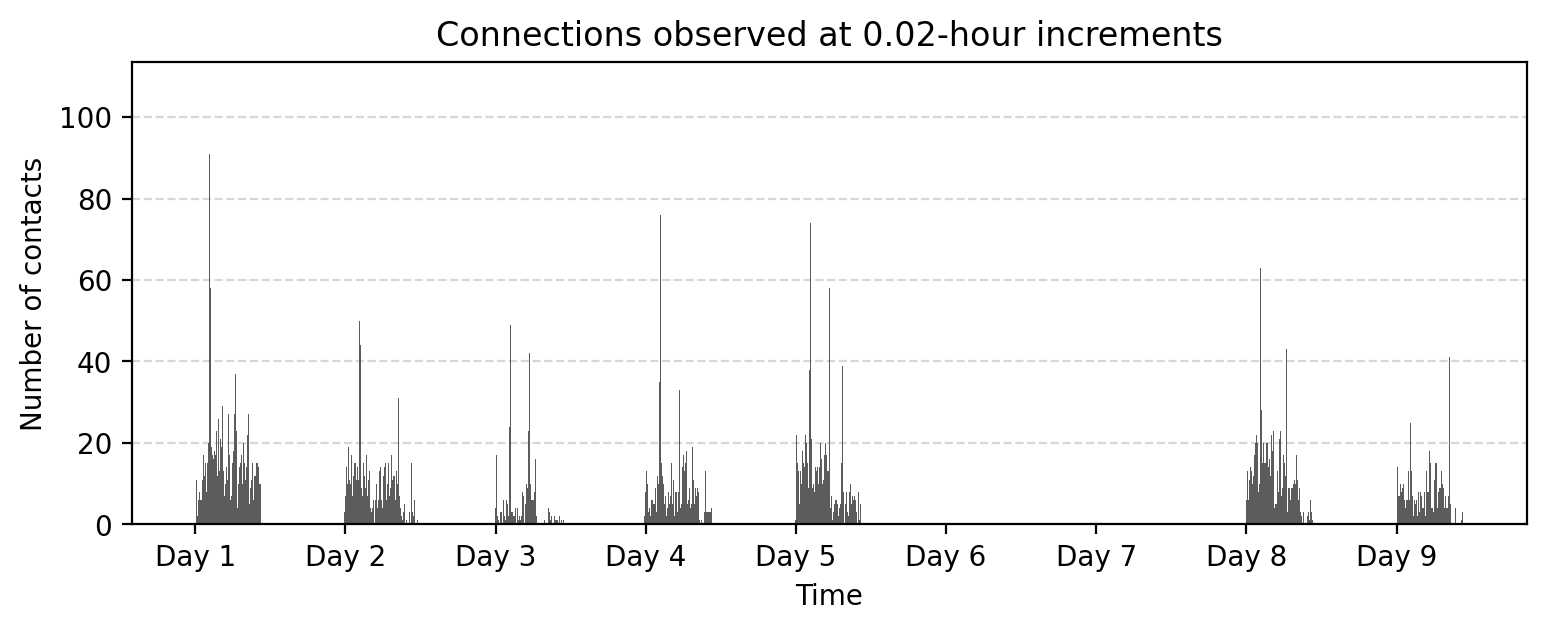
fig, ax = plt.subplots(1,1,figsize=(9,3),dpi=200)
ax.bar(aggregate_counts.index, aggregate_counts.values, width=bin_size, color='.2', alpha=0.8)
ax.grid(axis="y", linestyle="--", alpha=0.5)
# Set x-ticks at absolute 24-hour intervals
start_time = aggregate_counts.index.min()
end_time = aggregate_counts.index.max()
# Generate 24-hour spaced timestamps
tick_positions = range(start_time, end_time + 1, 24 * 3600) # Step by 24 hours (in seconds)
tick_labels = ['Day %i'%i for i in range(1,len(tick_positions)+1)]
# Set ticks and labels
ax.set_xticks(tick_positions)
ax.set_xticklabels(tick_labels)
ax.set_title("Connections observed at %.2f-hour increments"%(bin_size/(3600)))
ax.set_xlabel("Time")
ax.set_ylabel("Number of contacts")
plt.show()
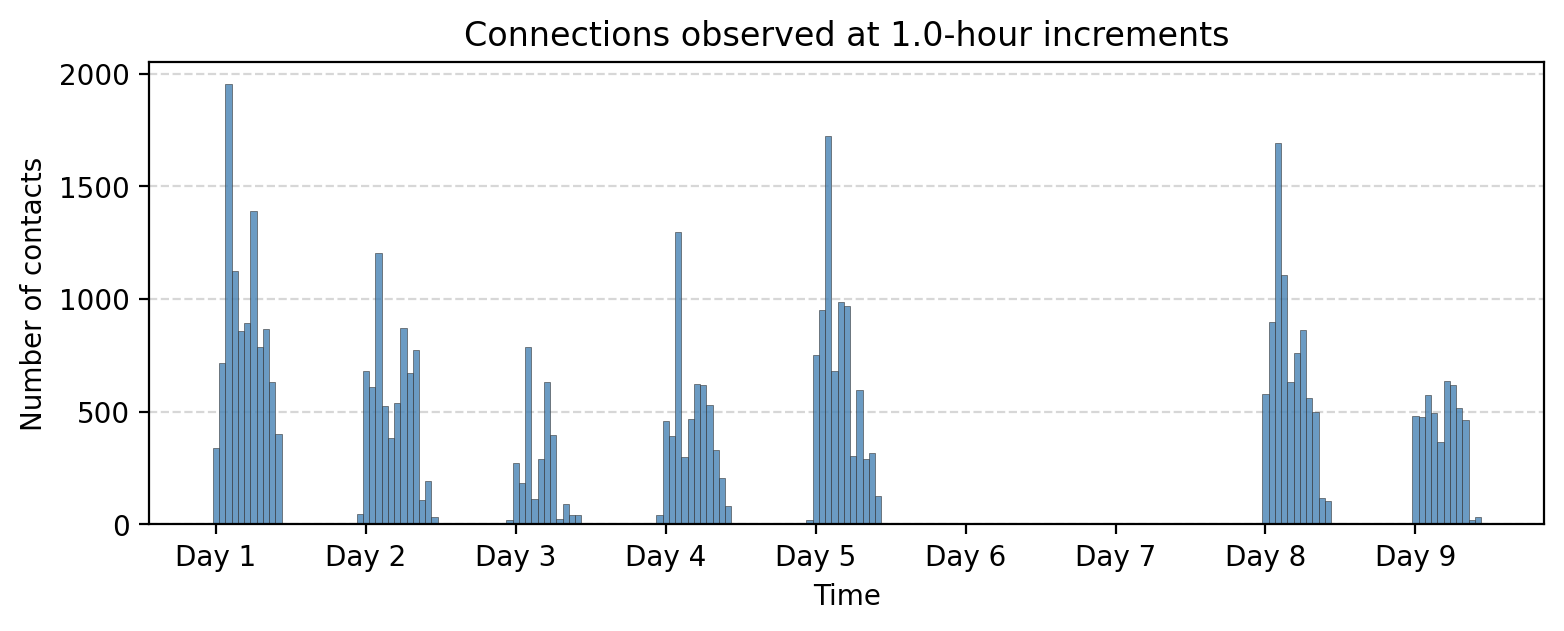
bin_size = 60*60 # 1 hour
tdf["time_bin"] = (tdf['timestamp'] // bin_size) * bin_size
aggregate_counts = tdf["time_bin"].value_counts().sort_index()
fig, ax = plt.subplots(1,1,figsize=(9,3),dpi=200)
ax.bar(aggregate_counts.index, aggregate_counts.values,
width=bin_size, color='steelblue', alpha=0.8, ec='.2', lw=0.25)
ax.grid(axis="y", linestyle="--", alpha=0.5)
# Set x-ticks at absolute 24-hour intervals
start_time = aggregate_counts.index.min()
end_time = aggregate_counts.index.max()
# Generate 24-hour spaced timestamps
tick_positions = range(start_time, end_time + 1, 24 * 3600) # Step by 24 hours (in seconds)
tick_labels = ['Day %i'%i for i in range(1,len(tick_positions)+1)]
# Set ticks and labels
ax.set_xticks(tick_positions)
ax.set_xticklabels(tick_labels)
ax.set_title("Connections observed at %.1f-hour increments"%(bin_size/(3600)))
ax.set_xlabel("Time")
ax.set_ylabel("Number of contacts")
plt.show()
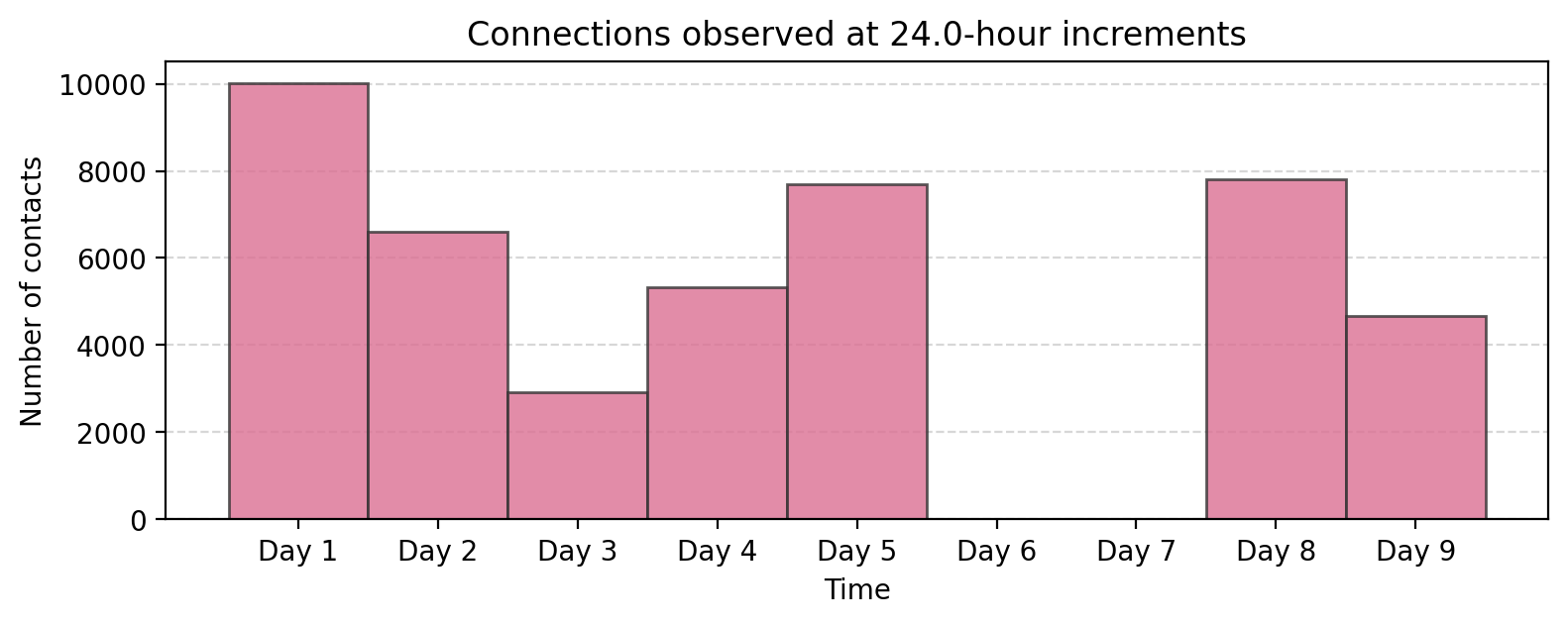
bin_size = 60*60*24 # 24 hour
tdf["time_bin"] = (tdf['timestamp'] // bin_size) * bin_size
aggregate_counts = tdf["time_bin"].value_counts().sort_index()
fig, ax = plt.subplots(1,1,figsize=(9,3),dpi=200)
ax.bar(aggregate_counts.index, aggregate_counts.values, width=bin_size,
color='palevioletred', alpha=0.8, ec='.2')
ax.grid(axis="y", linestyle="--", alpha=0.5)
# Set x-ticks at absolute 24-hour intervals
start_time = aggregate_counts.index.min()
end_time = aggregate_counts.index.max()
# Generate 24-hour spaced timestamps
tick_positions = range(start_time, end_time + 1, 24 * 3600) # Step by 24 hours (in seconds)
tick_labels = ['Day %i'%i for i in range(1,len(tick_positions)+1)]
# Set ticks and labels
ax.set_xticks(tick_positions)
ax.set_xticklabels(tick_labels)
ax.set_title("Connections observed at %.1f-hour increments"%(bin_size/(3600)))
ax.set_xlabel("Time")
ax.set_ylabel("Number of contacts")
plt.show()
But what about the network!#
def temporal_network_from_data(
data,
aggregation="all",
interval_size=1,
time_window=None,
custom_slices=None,
directed=False,
multigraph=False,
column_names=None
):
"""
Convert temporal edge data into NetworkX graph objects with flexible temporal aggregation.
This function processes temporal edge data (provided as a DataFrame, CSV file, or NumPy array)
to create NetworkX graphs. It supports various temporal aggregation methods, including grouping
by fixed intervals, custom time slices, or a single aggregated graph for all data.
Parameters:
----------
data : pd.DataFrame, str, or np.ndarray
Input temporal edge data. Can be:
- A Pandas DataFrame with required columns: 'node_i', 'node_j', 'timestamp'.
- A file path to a CSV file (requires `column_names` to be specified).
- A NumPy array with columns matching the required structure (requires `column_names`).
aggregation : str, optional
Temporal aggregation method. Options are:
- 'interval': Group data into fixed time intervals (specified by `interval_size`).
- 'custom': Group data based on custom time slices (specified by `custom_slices`).
- 'all': Aggregate all data into a single graph.
Default is 'all'.
interval_size : int, optional
Size of the time intervals (in seconds) for 'interval' aggregation. Default is 1.
time_window : tuple, optional
A tuple `(start, stop)` to filter timestamps within a specific range (in seconds).
Default is None, which includes all timestamps.
custom_slices : list, optional
List of custom slice sizes (in seconds) for 'custom' aggregation. For example, [3600, 7200]
would create two time bins of 1 hour and 2 hours, respectively. Default is None.
directed : bool, optional
If True, creates directed graphs (DiGraph or MultiDiGraph). Default is False.
multigraph : bool, optional
If True, creates MultiGraphs or MultiDiGraphs to allow parallel edges. If False,
standard Graph or DiGraph objects are created, and weights are aggregated. Default is False.
column_names : list, optional
Required when `data` is a CSV file or NumPy array. Specifies the column names as:
['node_i', 'node_j', 'timestamp', (optional) 'weight'].
Returns:
-------
dict or networkx.Graph
- If data is aggregated into multiple time bins (e.g., 'interval' or 'custom'), returns a
dictionary where keys are time bins and values are NetworkX graph objects.
- If data is aggregated into a single graph (e.g., 'all'), returns a single NetworkX graph.
Raises:
------
ValueError
- If required columns ('node_i', 'node_j', 'timestamp') are missing.
- If `custom_slices` is not provided for 'custom' aggregation.
- If `column_names` is not provided when input data is a CSV file or NumPy array.
TypeError
- If input data is not a Pandas DataFrame, a CSV file path, or a NumPy array.
Examples:
--------
1. Aggregating into a single graph:
>>> G_agg = temporal_network_from_data(data, aggregation="all")
2. Aggregating into 1-hour intervals:
>>> G_interval = temporal_network_from_data(data, aggregation="interval", interval_size=3600)
3. Using custom time slices:
>>> G_custom = temporal_network_from_data(data, aggregation="custom", custom_slices=[3600, 7200])
4. Handling a CSV file:
>>> G_csv = temporal_network_from_data("edges.csv",
column_names=["node_i","node_j","timestamp","weight"])
Notes:
-----
- The 'weight' column, if present, is aggregated for standard graphs (not MultiGraphs).
- The timestamp column must be numeric and represents time in seconds.
"""
# Handle data input
if isinstance(data, pd.DataFrame):
df = data
elif isinstance(data, str): # CSV file path
if column_names is None:
raise ValueError("Column names must be provided when reading from a CSV file.")
df = pd.read_csv(data, names=column_names)
elif isinstance(data, np.ndarray): # NumPy array
if column_names is None:
raise ValueError("Column names must be provided for a NumPy array.")
df = pd.DataFrame(data, columns=column_names)
else:
raise TypeError("Input data must be a Pandas DataFrame, a CSV file path, or a NumPy array.")
# Check required columns
required_columns = {"node_i", "node_j", "timestamp"}
if not required_columns.issubset(df.columns):
raise ValueError(f"Input data must contain {required_columns} columns.")
# Filter by time window
if time_window:
start, stop = time_window
df = df[(df["timestamp"] >= start) & (df["timestamp"] <= stop)]
# Determine time bins
if aggregation == "interval":
df["time_bin"] = (df["timestamp"] // interval_size) * interval_size
elif aggregation == "custom":
if not custom_slices:
raise ValueError("Custom slices must be provided for 'custom' aggregation.")
time_bins = []
current_start = df["timestamp"].min()
for slice_size in custom_slices:
current_end = current_start + slice_size
time_bins.append((current_start, current_end))
current_start = current_end
# Assign rows to custom time bins
def assign_to_custom_bin(ts):
for i, (start, end) in enumerate(time_bins):
if start <= ts < end:
return i
return None # Outside custom slices
df["time_bin"] = df["timestamp"].apply(assign_to_custom_bin)
df = df[df["time_bin"].notnull()] # Remove rows outside custom slices
elif aggregation == "all":
df["time_bin"] = "all" # Single graph for all data
else:
raise ValueError(f"Unsupported aggregation type: {aggregation}")
# Group by time bins and create graphs
graphs = {}
for time_bin, group in df.groupby("time_bin"):
# Determine graph type
if multigraph:
G = nx.MultiDiGraph() if directed else nx.MultiGraph()
else:
G = nx.DiGraph() if directed else nx.Graph()
# Aggregate weights if multigraph is False
if not multigraph:
edge_weights = (
group.groupby(["node_i", "node_j"])
.agg(weight=("weight", "sum") if "weight" in group.columns else ("timestamp", "size"))
.reset_index()
)
for _, row in edge_weights.iterrows():
G.add_edge(row["node_i"], row["node_j"], weight=row["weight"])
else: # Add edges directly without aggregation
for _, row in group.iterrows():
G.add_edge(
row["node_i"],
row["node_j"],
weight=row.get("weight", 1),
timestamp=row["timestamp"]
)
graphs[time_bin] = G
# Return a single graph if no aggregation
if aggregation == "all" or (aggregation == "custom" and len(graphs) == 1):
return graphs["all"]
return graphs
Test out our temporal network creation function! Visualize the aggregated network.
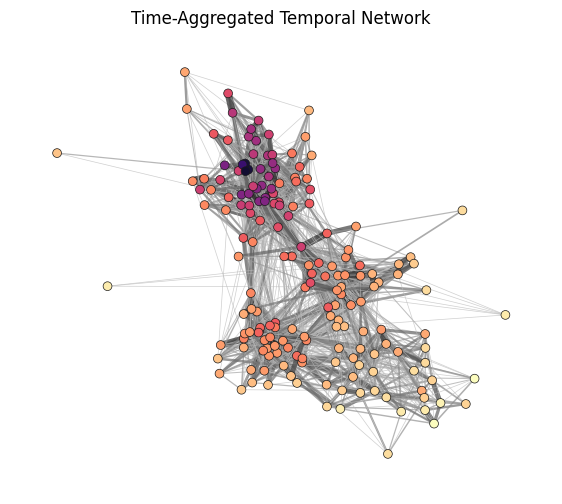
G_agg = temporal_network_from_data(tdf, aggregation="all")
np.random.seed(5)
pos = nx.spring_layout(G_agg, k=1.0, iterations=200, threshold=0.005)
fig, ax = plt.subplots(1, 1, figsize=(7,6), dpi=100)
# Extract edge weights
ews = np.array(list(nx.get_edge_attributes(G_agg, 'weight').values()))
ews = ews ** 0.3 - 0.5 # Scale edge width non-linearly for visualization
# Relate edge colors to the normalized edge width and adjust with an exponent
ecs = (ews / max(ews)) ** 0.15
# Calculate eigenvector centrality for nodes, adjust with an exponent for better visualization
ncs = np.array(list(nx.eigenvector_centrality(G_agg, weight='weight').values())) ** 0.1
ncs = (ncs-min(ncs))/(max(ncs)-min(ncs))
# Draw the nodes of the network
nx.draw_networkx_nodes(
G_agg, # The aggregated temporal network graph
pos, # The positions of nodes (should be precomputed separately)
node_color=ncs, # Color nodes based on their eigenvector centrality scores
node_size=40, # Size of the nodes
linewidths=0.5, # Border width of nodes
edgecolors='.1', # Edge color of the nodes (dark grey)
cmap=plt.cm.magma_r, # Node colormap
vmin=0, vmax=1.1,
ax=ax
)
nx.draw_networkx_edges(
G_agg, # The aggregated temporal network graph
pos, # The positions of nodes
width=ews, # Edge widths calculated above
edge_color=ecs, # Edge colors calculated above
edge_cmap=plt.cm.Greys, # Use the Greys colormap for edges
edge_vmin=0.45, # Minimum value for edge color normalization
edge_vmax=1.0, # Maximum value for edge color normalization
alpha=0.7, # Transparency of edges
ax=ax
)
ax.set_title('Time-Aggregated Temporal Network')
ax.set_axis_off()
plt.show()
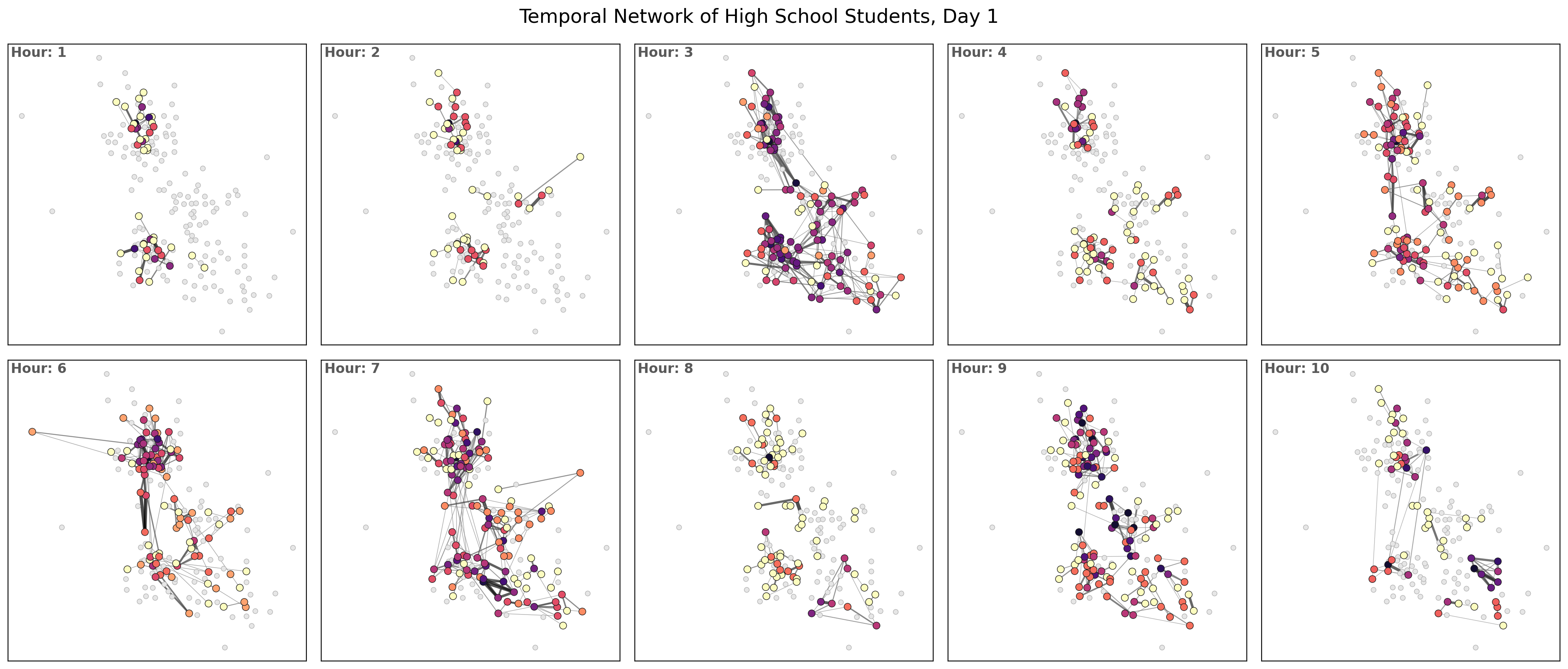
What about if we want a snapshot of the temporal network? E.g. the first hour?
day = 0
int_size = 60*60
G_t = temporal_network_from_data(tdf,
aggregation="interval", # Options: 'interval', 'custom', 'all'
interval_size=int_size, # Size of the time interval for 'interval' aggregation
time_window=None, # Tuple (start, stop) to restrict aggregation to specific time range
custom_slices=None, # List of custom slice sizes for aggregation
directed=False, # Create directed graph
multigraph=False, # Create a MultiGraph or MultiDiGraph instead of Graph/DiGraph
column_names=None) # Required if `data` is a CSV or NumPy array
G_t
{0: <networkx.classes.graph.Graph at 0x7f05795bfcd0>,
3600: <networkx.classes.graph.Graph at 0x7f058c953dd0>,
7200: <networkx.classes.graph.Graph at 0x7f05798359d0>,
10800: <networkx.classes.graph.Graph at 0x7f057975ba90>,
14400: <networkx.classes.graph.Graph at 0x7f0579822750>,
18000: <networkx.classes.graph.Graph at 0x7f05795bfb10>,
21600: <networkx.classes.graph.Graph at 0x7f057974ccd0>,
25200: <networkx.classes.graph.Graph at 0x7f058c9afc90>,
28800: <networkx.classes.graph.Graph at 0x7f057975a490>,
32400: <networkx.classes.graph.Graph at 0x7f0579a9f210>,
36000: <networkx.classes.graph.Graph at 0x7f05796cde10>,
82800: <networkx.classes.graph.Graph at 0x7f057999a810>,
86400: <networkx.classes.graph.Graph at 0x7f05796f3010>,
90000: <networkx.classes.graph.Graph at 0x7f0579996fd0>,
93600: <networkx.classes.graph.Graph at 0x7f057da92f90>,
97200: <networkx.classes.graph.Graph at 0x7f0576046bd0>,
100800: <networkx.classes.graph.Graph at 0x7f057974e3d0>,
104400: <networkx.classes.graph.Graph at 0x7f05798adb50>,
108000: <networkx.classes.graph.Graph at 0x7f05760ae110>,
111600: <networkx.classes.graph.Graph at 0x7f0579a8d890>,
115200: <networkx.classes.graph.Graph at 0x7f05760c8910>,
118800: <networkx.classes.graph.Graph at 0x7f057973fc90>,
122400: <networkx.classes.graph.Graph at 0x7f05796d5390>,
126000: <networkx.classes.graph.Graph at 0x7f0576085350>,
169200: <networkx.classes.graph.Graph at 0x7f05760c16d0>,
172800: <networkx.classes.graph.Graph at 0x7f0579851890>,
176400: <networkx.classes.graph.Graph at 0x7f05760e6250>,
180000: <networkx.classes.graph.Graph at 0x7f05796ee550>,
183600: <networkx.classes.graph.Graph at 0x7f057983bb50>,
187200: <networkx.classes.graph.Graph at 0x7f0579879610>,
190800: <networkx.classes.graph.Graph at 0x7f05760a3750>,
194400: <networkx.classes.graph.Graph at 0x7f057974e810>,
198000: <networkx.classes.graph.Graph at 0x7f0575f3b490>,
201600: <networkx.classes.graph.Graph at 0x7f05760847d0>,
205200: <networkx.classes.graph.Graph at 0x7f0576093690>,
208800: <networkx.classes.graph.Graph at 0x7f0576092bd0>,
255600: <networkx.classes.graph.Graph at 0x7f0576061f10>,
259200: <networkx.classes.graph.Graph at 0x7f0575f3b750>,
262800: <networkx.classes.graph.Graph at 0x7f057981e610>,
266400: <networkx.classes.graph.Graph at 0x7f0579aaf590>,
270000: <networkx.classes.graph.Graph at 0x7f0576092a50>,
273600: <networkx.classes.graph.Graph at 0x7f0575f82790>,
277200: <networkx.classes.graph.Graph at 0x7f0575f49850>,
280800: <networkx.classes.graph.Graph at 0x7f0575f52c10>,
284400: <networkx.classes.graph.Graph at 0x7f0575fcb7d0>,
288000: <networkx.classes.graph.Graph at 0x7f0575f3be50>,
291600: <networkx.classes.graph.Graph at 0x7f05795a1ad0>,
295200: <networkx.classes.graph.Graph at 0x7f0575fcb850>,
298800: <networkx.classes.graph.Graph at 0x7f0579aca650>,
302400: <networkx.classes.graph.Graph at 0x7f0575f3ac50>,
342000: <networkx.classes.graph.Graph at 0x7f057987bad0>,
345600: <networkx.classes.graph.Graph at 0x7f05760a2fd0>,
349200: <networkx.classes.graph.Graph at 0x7f05760625d0>,
352800: <networkx.classes.graph.Graph at 0x7f0575f84850>,
356400: <networkx.classes.graph.Graph at 0x7f0579996c50>,
360000: <networkx.classes.graph.Graph at 0x7f05760a31d0>,
363600: <networkx.classes.graph.Graph at 0x7f05796f3190>,
367200: <networkx.classes.graph.Graph at 0x7f0576004650>,
370800: <networkx.classes.graph.Graph at 0x7f057984a990>,
374400: <networkx.classes.graph.Graph at 0x7f0575ffb5d0>,
378000: <networkx.classes.graph.Graph at 0x7f057603cb90>,
381600: <networkx.classes.graph.Graph at 0x7f0575f972d0>,
601200: <networkx.classes.graph.Graph at 0x7f057611d250>,
604800: <networkx.classes.graph.Graph at 0x7f05760e5bd0>,
608400: <networkx.classes.graph.Graph at 0x7f05796cf110>,
612000: <networkx.classes.graph.Graph at 0x7f0575e6ba10>,
615600: <networkx.classes.graph.Graph at 0x7f0575e8ff90>,
619200: <networkx.classes.graph.Graph at 0x7f057974cc50>,
622800: <networkx.classes.graph.Graph at 0x7f0575f3b7d0>,
626400: <networkx.classes.graph.Graph at 0x7f0575fb9d90>,
630000: <networkx.classes.graph.Graph at 0x7f0575fc5ed0>,
633600: <networkx.classes.graph.Graph at 0x7f057973e0d0>,
637200: <networkx.classes.graph.Graph at 0x7f05760fe690>,
640800: <networkx.classes.graph.Graph at 0x7f05760c8410>,
687600: <networkx.classes.graph.Graph at 0x7f0575fab6d0>,
691200: <networkx.classes.graph.Graph at 0x7f05760d9cd0>,
694800: <networkx.classes.graph.Graph at 0x7f057603c4d0>,
698400: <networkx.classes.graph.Graph at 0x7f0575e86cd0>,
702000: <networkx.classes.graph.Graph at 0x7f05799dfe50>,
705600: <networkx.classes.graph.Graph at 0x7f0575f1be50>,
709200: <networkx.classes.graph.Graph at 0x7f05799a8850>,
712800: <networkx.classes.graph.Graph at 0x7f0575e872d0>,
716400: <networkx.classes.graph.Graph at 0x7f057986c410>,
720000: <networkx.classes.graph.Graph at 0x7f057974fbd0>,
723600: <networkx.classes.graph.Graph at 0x7f0575feb510>,
727200: <networkx.classes.graph.Graph at 0x7f05761229d0>}
import itertools as it
nrows = 2
ncols = 5
hours_to_plot = nrows*ncols
fig, ax = plt.subplots(nrows, ncols, figsize=(5*ncols,5*nrows), dpi=200)
tups = list(it.product(range(nrows), range(ncols)))
plt.subplots_adjust(wspace=0.05, hspace=0.05)
for ti, t in enumerate(list(G_t.keys())[:hours_to_plot]):
a = tups[ti]
Gt = G_t[t]
nx.draw_networkx_nodes(G_agg, pos, node_color='.9',
node_size=20, linewidths=0.5, edgecolors='.7', ax=ax[a])
ews = np.array(list(nx.get_edge_attributes(Gt, 'weight').values()))
ews = ews ** 0.3 - 0.5 # Scale edge width non-linearly for visualization
ecs = (ews / max(ews)) ** 0.15
ncs = np.array(list(nx.degree_centrality(Gt).values())) ** 0.1
ncs = (ncs-min(ncs))/(max(ncs)-min(ncs))
nx.draw_networkx_nodes(Gt, pos, node_color=ncs, node_size=40, linewidths=0.5,
edgecolors='.1', cmap=plt.cm.magma_r, vmin=0, vmax=1.1, ax=ax[a])
nx.draw_networkx_edges(Gt, pos, width=ews, edge_color=ecs, edge_cmap=plt.cm.Greys,
edge_vmin=0.45, edge_vmax=1.0, alpha=0.7, ax=ax[a])
ax[a].set_title('Hour: %i'%(ti+1), ha='left', va='top', x=0.01,
fontweight='bold', y=0.97, color='.35')
plt.suptitle('Temporal Network of High School Students, Day %i'%(day+1), y=0.925, fontsize='xx-large')
plt.show()
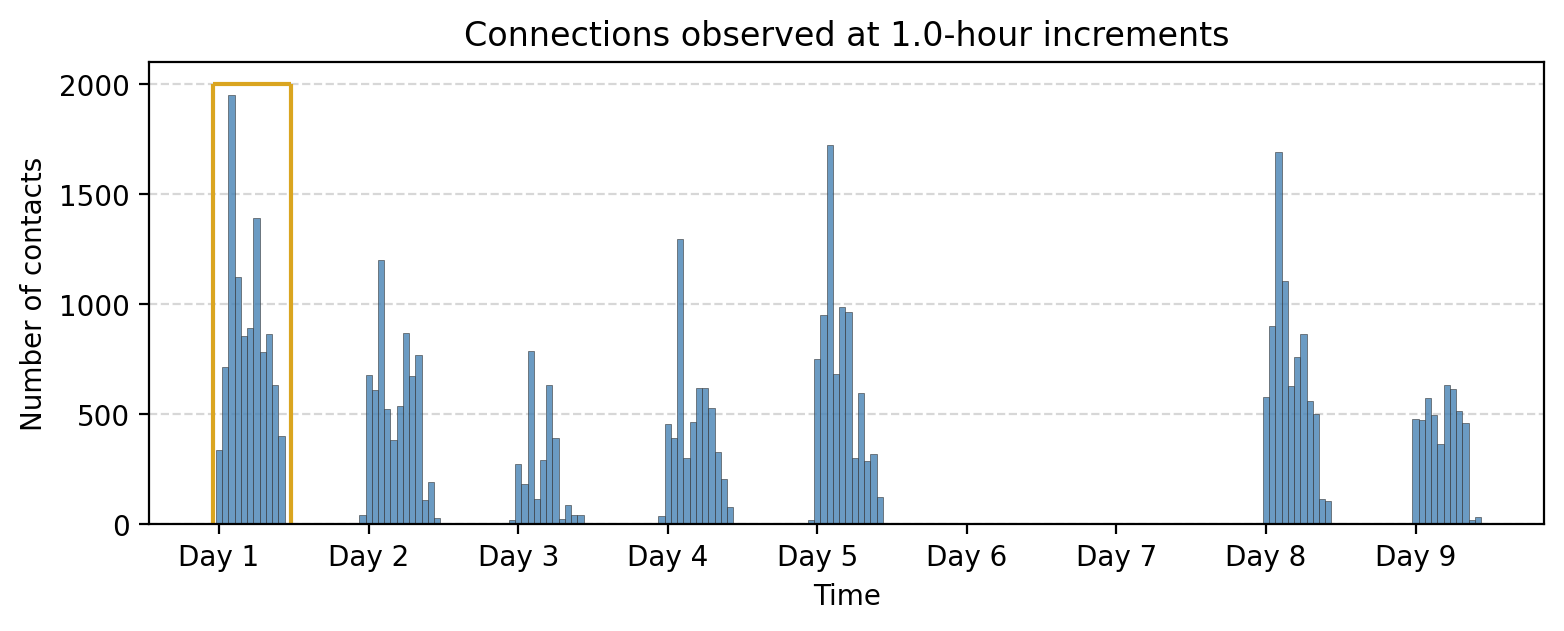
bin_size = 60*60 # 1 hour
tdf["time_bin"] = (tdf['timestamp'] // bin_size) * bin_size
aggregate_counts = tdf["time_bin"].value_counts().sort_index()
fig, ax = plt.subplots(1,1,figsize=(9,3),dpi=200)
ax.bar(aggregate_counts.index, aggregate_counts.values,
width=bin_size, color='steelblue', alpha=0.8, ec='.2', lw=0.25)
ax.vlines(-60*60, 0, 2000, color='goldenrod')
ax.hlines(2000, -60*60, 60*60*11.5, color='goldenrod')
ax.vlines(60*60*11.5, 0, 2000, color='goldenrod')
ax.grid(axis="y", linestyle="--", alpha=0.5)
# Set x-ticks at absolute 24-hour intervals
start_time = aggregate_counts.index.min()
end_time = aggregate_counts.index.max()
# Generate 24-hour spaced timestamps
tick_positions = range(start_time, end_time + 1, 24 * 3600) # Step by 24 hours (in seconds)
tick_labels = ['Day %i'%i for i in range(1,len(tick_positions)+1)]
# Set ticks and labels
ax.set_xticks(tick_positions)
ax.set_xticklabels(tick_labels)
ax.set_title("Connections observed at %.1f-hour increments"%(bin_size/(3600)))
ax.set_xlabel("Time")
ax.set_ylabel("Number of contacts")
plt.show()
def get_binning(data, num_bins=50, is_pmf=False, log_binning=False, threshold=0):
"""
Bins the input data and calculates the probability mass function (PMF) or
probability density function (PDF) over the bins. Supports both linear and
logarithmic binning.
Parameters
----------
data : array-like
The data to be binned, typically a list or numpy array of values.
num_bins : int, optional
The number of bins to use for binning the data (default is 15).
is_pmf : bool, optional
If True, computes the probability mass function (PMF) by normalizing
histogram counts to sum to 1. If False, computes the probability density
function (PDF) by normalizing the density of the bins (default is True).
log_binning : bool, optional
If True, uses logarithmic binning with log-spaced bins. If False, uses
linear binning (default is False).
threshold : float, optional
Only values greater than `threshold` will be included in the binning,
allowing for the removal of isolated nodes or outliers (default is 0).
Returns
-------
x : numpy.ndarray
The bin centers, adjusted to be the midpoint of each bin.
p : numpy.ndarray
The computed PMF or PDF values for each bin.
Notes
-----
This function removes values below a specified threshold, then defines
bin edges based on the specified binning method (linear or logarithmic).
It calculates either the PMF or PDF based on `is_pmf`.
"""
# Filter out isolated nodes or low values by removing data below threshold
values = list(filter(lambda x: x > threshold, data))
if len(values) != len(data):
print("%s isolated nodes have been removed" % (len(data) - len(values)))
# Define the range for binning (support of the distribution)
lower_bound = min(values)
upper_bound = max(values)
# Define bin edges based on binning type (logarithmic or linear)
if log_binning:
# Use log-spaced bins by taking the log of the bounds
lower_bound = np.log10(lower_bound)
upper_bound = np.log10(upper_bound)
bin_edges = np.logspace(lower_bound, upper_bound, num_bins + 1, base=10)
else:
# Use linearly spaced bins
bin_edges = np.linspace(lower_bound, upper_bound, num_bins + 1)
# Calculate histogram based on chosen binning method
if is_pmf:
# Calculate PMF: normalized counts of data in each bin
y, _ = np.histogram(values, bins=bin_edges, density=False)
p = y / y.sum() # Normalize to get probabilities
else:
# Calculate PDF: normalized density of data in each bin
p, _ = np.histogram(values, bins=bin_edges, density=True)
# Compute bin centers (midpoints) to represent each bin
x = bin_edges[1:] - np.diff(bin_edges) / 2 # Bin centers for plotting
# Remove bins with zero probability to avoid plotting/display issues
x = x[p > 0]
p = p[p > 0]
return x, p
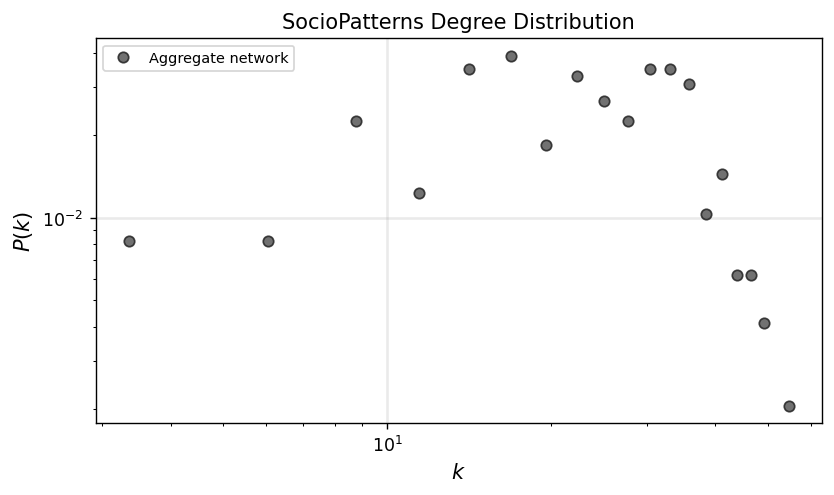
degrees = np.array(list(dict(G_agg.degree()).values()))
x_agg, y_agg = get_binning(degrees, num_bins=20)
fig, ax = plt.subplots(1,1,figsize=(7.5,4),dpi=125)
ax.loglog(x_agg, y_agg, 'o', color='.3', label='Aggregate network', alpha=0.8, mec='.1')
ax.set_xlabel(r"$k$",fontsize='large')
ax.set_ylabel(r"$P(k)$",fontsize='large')
ax.legend(fontsize='small')
ax.grid(linewidth=1.5, color='#999999', alpha=0.2, linestyle='-')
ax.set_title('SocioPatterns Degree Distribution')
plt.show()
interval_sizes = [86400, 172800, 259200, 345600, 432000, 518400, 604800, 691200, 777600]
int_s = interval_sizes[0]
G_t = temporal_network_from_data(tdf,
aggregation="interval", # Options: 'interval', 'custom', 'all'
interval_size=int_s, # Size of the time interval for 'interval' aggregation
time_window=None, # Tuple (start, stop) to restrict aggregation to specific time range
custom_slices=None, # List of custom slice sizes for aggregation
directed=False, # Create directed graph
multigraph=False, # Create a MultiGraph or MultiDiGraph instead of Graph/DiGraph
column_names=None) # Required if `data` is a CSV or NumPy array
Gt_i = G_t[0]
degrees = np.array(list(dict(G_agg.degree()).values()))
x_agg, y_agg = get_binning(degrees, num_bins=20)
weights_agg = np.array(list(nx.get_edge_attributes(G_agg, 'weight').values()))
xw_agg, yw_agg = get_binning(weights_agg, num_bins=20)
fig, ax = plt.subplots(1,2,figsize=(12,4),dpi=125)
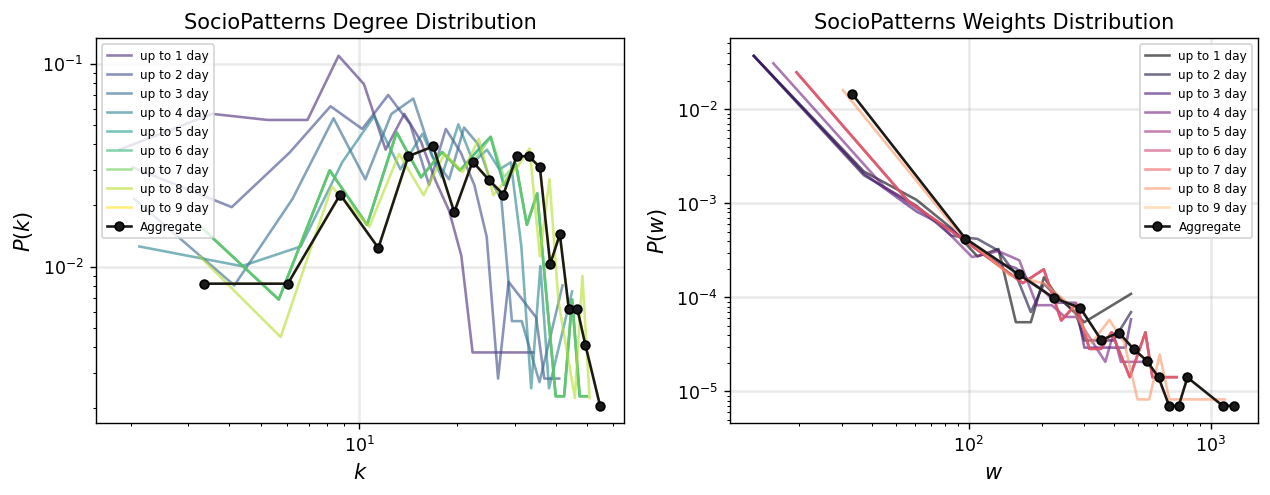
interval_sizes = [86400, 172800, 259200, 345600, 432000, 518400, 604800, 691200, 777600]
cols = plt.cm.viridis(np.linspace(0,1,1+len(interval_sizes)))[1:]
colsw = plt.cm.magma(np.linspace(0,1,1+len(interval_sizes)))[:-1]
for i,int_s in enumerate(interval_sizes):
Gt_i = temporal_network_from_data(tdf, aggregation="interval", interval_size=int_s)[0]
degrees_ti = np.array(list(dict(Gt_i.degree()).values()))
x_ti, y_ti = get_binning(degrees_ti, num_bins=20)
ax[0].loglog(x_ti, y_ti, '-', color=cols[i], label='up to %i day'%(i+1),
alpha=0.6)
weights_ti = np.array(list(nx.get_edge_attributes(Gt_i, 'weight').values()))
xw_ti, yw_ti = get_binning(weights_ti, num_bins=20)
ax[1].loglog(xw_ti, yw_ti, '-', color=colsw[i], label='up to %i day'%(i+1),
alpha=0.6)
ax[0].loglog(x_agg, y_agg, '.-', color='.1', label='Aggregate', mec='.0', ms=10)
ax[1].loglog(xw_agg, yw_agg, '.-', color='.1', label='Aggregate', mec='.0', ms=10)
ax[0].set_xlabel(r"$k$",fontsize='large')
ax[0].set_ylabel(r"$P(k)$",fontsize='large')
ax[0].legend(fontsize='x-small',loc=2)
ax[0].grid(linewidth=1.5, color='#999999', alpha=0.2, linestyle='-')
ax[0].set_title('SocioPatterns Degree Distribution')
ax[1].set_xlabel(r"$w$",fontsize='large')
ax[1].set_ylabel(r"$P(w)$",fontsize='large')
ax[1].legend(fontsize='x-small',loc=1)
ax[1].grid(linewidth=1.5, color='#999999', alpha=0.2, linestyle='-')
ax[1].set_title('SocioPatterns Weights Distribution')
plt.show()
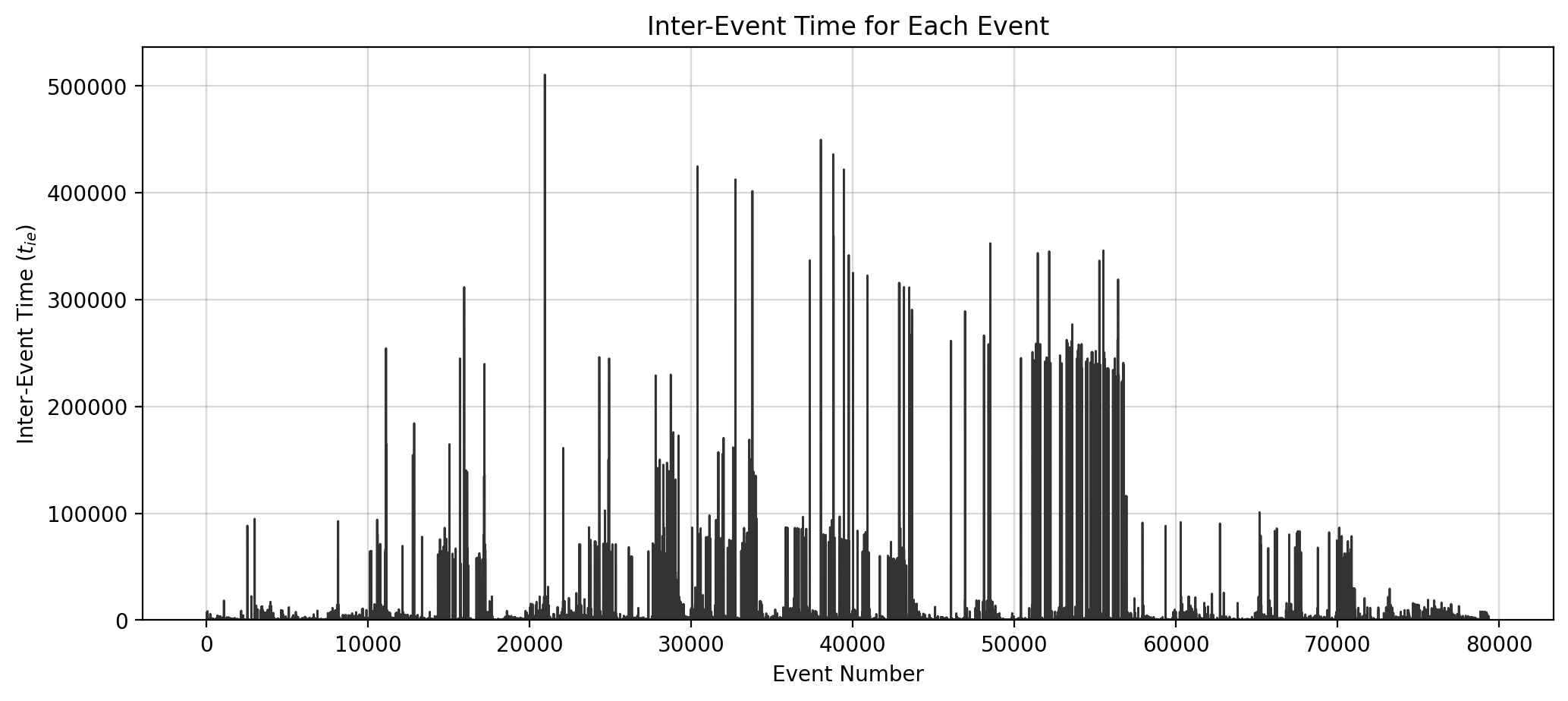
t_net = temporal_network_from_data(tdf, aggregation='interval')
---------------------------------------------------------------------------
KeyboardInterrupt Traceback (most recent call last)
Cell In[23], line 1
----> 1 t_net = temporal_network_from_data(tdf, aggregation='interval')
Cell In[11], line 157, in temporal_network_from_data(data, aggregation, interval_size, time_window, custom_slices, directed, multigraph, column_names)
153 # Aggregate weights if multigraph is False
154 if not multigraph:
155 edge_weights = (
156 group.groupby(["node_i", "node_j"])
--> 157 .agg(weight=("weight", "sum") if "weight" in group.columns else ("timestamp", "size"))
158 .reset_index()
159 )
160 for _, row in edge_weights.iterrows():
161 G.add_edge(row["node_i"], row["node_j"], weight=row["weight"])
File /opt/hostedtoolcache/Python/3.11.13/x64/lib/python3.11/site-packages/pandas/core/groupby/generic.py:902, in DataFrameGroupBy.aggregate(self, func, engine, engine_kwargs, *args, **kwargs)
898 elif relabeling and result is not None:
899 # this should be the only (non-raising) case with relabeling
900 # used reordered index of columns
901 result = result.iloc[:, order]
--> 902 result.columns = columns
904 if result is None:
905
906 # grouper specific aggregations
907 if self.grouper.nkeys > 1:
908 # test_groupby_as_index_series_scalar gets here with 'not self.as_index'
File /opt/hostedtoolcache/Python/3.11.13/x64/lib/python3.11/site-packages/pandas/core/generic.py:5915, in NDFrame.__setattr__(self, name, value)
5913 try:
5914 object.__getattribute__(self, name)
-> 5915 return object.__setattr__(self, name, value)
5916 except AttributeError:
5917 pass
File /opt/hostedtoolcache/Python/3.11.13/x64/lib/python3.11/site-packages/pandas/_libs/properties.pyx:69, in pandas._libs.properties.AxisProperty.__set__()
File /opt/hostedtoolcache/Python/3.11.13/x64/lib/python3.11/site-packages/pandas/core/generic.py:822, in NDFrame._set_axis(self, axis, labels)
821 def _set_axis(self, axis: int, labels: AnyArrayLike | list) -> None:
--> 822 labels = ensure_index(labels)
823 self._mgr.set_axis(axis, labels)
824 self._clear_item_cache()
File /opt/hostedtoolcache/Python/3.11.13/x64/lib/python3.11/site-packages/pandas/core/indexes/base.py:7333, in ensure_index(index_like, copy)
7331 return Index._with_infer(index_like, copy=copy, tupleize_cols=False)
7332 else:
-> 7333 return Index._with_infer(index_like, copy=copy)
File /opt/hostedtoolcache/Python/3.11.13/x64/lib/python3.11/site-packages/pandas/core/indexes/base.py:716, in Index._with_infer(cls, *args, **kwargs)
714 with warnings.catch_warnings():
715 warnings.filterwarnings("ignore", ".*the Index constructor", FutureWarning)
--> 716 result = cls(*args, **kwargs)
718 if result.dtype == _dtype_obj and not result._is_multi:
719 # error: Argument 1 to "maybe_convert_objects" has incompatible type
720 # "Union[ExtensionArray, ndarray[Any, Any]]"; expected
721 # "ndarray[Any, Any]"
722 values = lib.maybe_convert_objects(result._values) # type: ignore[arg-type]
File /opt/hostedtoolcache/Python/3.11.13/x64/lib/python3.11/site-packages/pandas/core/indexes/base.py:568, in Index.__new__(cls, data, dtype, copy, name, tupleize_cols, **kwargs)
565 subarr = com.asarray_tuplesafe(data, dtype=_dtype_obj)
566 if dtype is None:
567 # with e.g. a list [1, 2, 3] casting to numeric is _not_ deprecated
--> 568 subarr = _maybe_cast_data_without_dtype(
569 subarr, cast_numeric_deprecated=False
570 )
571 dtype = subarr.dtype
572 return Index(subarr, dtype=dtype, copy=copy, name=name, **kwargs)
File /opt/hostedtoolcache/Python/3.11.13/x64/lib/python3.11/site-packages/pandas/core/indexes/base.py:7414, in _maybe_cast_data_without_dtype(subarr, cast_numeric_deprecated)
7396 def _maybe_cast_data_without_dtype(
7397 subarr: np.ndarray, cast_numeric_deprecated: bool = True
7398 ) -> ArrayLike:
7399 """
7400 If we have an arraylike input but no passed dtype, try to infer
7401 a supported dtype.
(...) 7411 np.ndarray or ExtensionArray
7412 """
-> 7414 result = lib.maybe_convert_objects(
7415 subarr,
7416 convert_datetime=True,
7417 convert_timedelta=True,
7418 convert_period=True,
7419 convert_interval=True,
7420 dtype_if_all_nat=np.dtype("datetime64[ns]"),
7421 )
7422 if result.dtype.kind in ["i", "u", "f"]:
7423 if not cast_numeric_deprecated:
7424 # i.e. we started with a list, not an ndarray[object]
KeyboardInterrupt:
nodes_events = dict()
for t in sorted(t_net.keys()): # Iterate over sorted time bins
graph = t_net[t]
for node in graph.nodes():
nodes_events.setdefault(node, []).append(t) # Track event times per node
# Step 2: Calculate inter-event times for each node
event_ie_t = dict()
for node, event_list in nodes_events.items(): # Iterate over nodes and their event times
for i in range(1, len(event_list)):
ie_t = event_list[i] - event_list[i - 1] # Calculate inter-event time
event_ie_t.setdefault(event_list[i - 1], []).append(ie_t) # Group by start time
# Step 3: Flatten the inter-event times for plotting
event_number = []
ie_time_list = []
counter = 0
for t in sorted(event_ie_t.keys()): # Iterate over sorted event start times
for ie_t in event_ie_t[t]:
event_number.append(counter) # Track event number
ie_time_list.append(ie_t) # Add inter-event time
counter += 1
fig, ax = plt.subplots(1,1,figsize=(12,5),dpi=200)
ax.plot(event_number, ie_time_list, color='.2', lw=1)
ax.set_xlabel("Event Number")
ax.set_ylabel("Inter-Event Time ($t_{ie}$)")
ax.set_title("Inter-Event Time for Each Event")
ax.grid(alpha=0.5)
ax.set_ylim(0)
# ax.set_xlim(0, 80000)
plt.show()
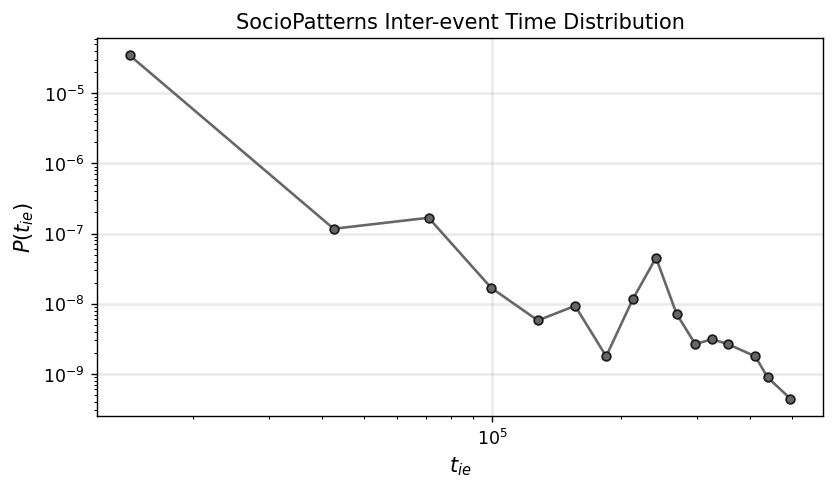
x_ie, y_ie = get_binning(ie_time_list, num_bins=18)
fig, ax = plt.subplots(1,1,figsize=(7.5,4),dpi=125)
ax.loglog(x_ie, y_ie, '.-', color='.4', mec='.1', ms=10)
ax.set_xlabel(r"$t_{ie}$",fontsize='large')
ax.set_ylabel(r"$P(t_{ie})$",fontsize='large')
ax.grid(linewidth=1.5, color='#999999', alpha=0.2, linestyle='-')
ax.set_title('SocioPatterns Inter-event Time Distribution')
plt.show()
Your turn!#
Select two global network properties to analyze over time. Pick a timescale to view your network evolution, and create a plot with two subplots that track this property over time.
pass
Background and Introduction to Burstiness Index for Temporal Networks#
Temporal networks are systems where interactions or connections between entities occur at specific times. Unlike static networks, temporal networks account for the timing and sequence of events, making them essential for understanding processes such as information diffusion, disease transmission, and human communication patterns.
One intriguing property of temporal networks is burstiness—the tendency of events to cluster in short periods of high activity, followed by long periods of inactivity. This phenomenon is observed across various domains, from human interactions (e.g., email exchanges, phone calls) to natural systems (e.g., neuronal firing, animal behavior).
Burstiness in Temporal Networks#
Burstiness arises when the timing of events deviates from a uniform or random (Poissonian) distribution. In a random process, inter-event times (the time gaps between consecutive events) are typically distributed exponentially. However, real-world systems often exhibit heavy-tailed distributions, where a significant fraction of inter-event times are very short, interspersed with much longer intervals.
This non-random clustering of events has significant implications:
Efficiency: Burstiness can accelerate or slow down dynamic processes like spreading phenomena.
Predictability: It reflects underlying mechanisms governing the system, such as behavioral patterns or network constraints.
System Design: Understanding burstiness helps optimize systems, from communication networks to epidemic models.
Burstiness Index#
The burstiness index is a quantitative measure introduced to capture this deviation from randomness in temporal event sequences. It is defined based on the statistical properties of inter-event times.
For a sequence of inter-event times \( \{ \tau_1, \tau_2, \dots, \tau_n \} \), the burstiness index \( B \) is defined as:
where:
\( \mu_\tau \) is the mean of the inter-event times,
\( \sigma_\tau \) is the standard deviation of the inter-event times.
The burstiness index \( B \) has the following properties:
\( B = -1 \): Events are perfectly regular (e.g., periodic).
\( B = 0 \): Events are Poissonian, with a random, exponential distribution of inter-event times.
\( B > 0 \): Events are bursty, with a heavy-tailed distribution of inter-event times.
Intuition Behind the Formula#
The burstiness index is designed to compare the variability (\( \sigma \)) of inter-event times to their mean (\( \mu \)):
When \( \sigma \approx \mu \), the inter-event times are relatively uniform, and \( B \) approaches 0.
When \( \sigma \gg \mu \), the variability dominates, indicating highly irregular event timing, and \( B \) becomes positive.
When \( \sigma \ll \mu \), as in periodic systems, \( B \) becomes negative.
Applications of Burstiness Index#
The burstiness index has been widely used to study temporal networks in various contexts:
Social Networks: Understanding the temporal patterns of human communication (e.g., email, messaging, phone calls).
Biological Systems: Analyzing neuronal firing patterns or animal behavior.
Infrastructure Networks: Characterizing traffic patterns or network usage in telecommunication systems.
Epidemiology: Modeling the timing of human interactions that drive disease spread.
Importance in Temporal Networks#
In temporal networks, burstiness provides critical insights into the structure and dynamics of event sequences. For example:
It influences how fast information or diseases propagate.
It reveals behavioral rhythms and anomalies in human activity.
It helps differentiate between natural and engineered systems.
The burstiness index is thus an essential tool for exploring and characterizing temporal networks, enabling researchers to identify deviations from randomness and uncover patterns underlying complex systems.
References
Barabási, AL. The origin of bursts and heavy tails in human dynamics. Nature 435, 207–211 (2005). https://doi.org/10.1038/nature03459
Goh, K. I., & Barabási, AL (2008). Burstiness and memory in complex systems. Europhysics Letters, 81(4), 48002. https://iopscience.iop.org/article/10.1209/0295-5075/81/48002
Karsai, M., Kaski, K., Barabási, AL. et al. Universal features of correlated bursty behaviour. Sci Rep 2, 397 (2012). https://doi.org/10.1038/srep00397
Origin of burstiness? Queue Model#
Possible causes? Heterogeneous distribution of priorities.
Priority List Model#
Pick the task with the highest priority with probability \( p \), or randomly pick one with \( 1-p \), and execute it.
Generate a new task with a random priority. | Task | Priority | |———-|————–| | 1 | \( x_1 \) | | 2 | \( x_2 \) | | 3 | \( x_3 \) | | 4 | \( x_4 \) | | 5 | \( x_5 \) | | 6 | \( x_6 \) | | … | … | | \( L \) | \( x_L \) |
The probability of executing a task is \(\Pi(x) = \dfrac{x^{\gamma}}{\sum_{i=1\cdots L} x_i^{\gamma}}\).
\(\gamma = 0\): random (\(p=0\))
\(\gamma = \infty\): determinsitic (\(p=1\))
The average waiting time of a task with priority \(x\) is executed \(\tau(x) = \dfrac{1}{\Pi(x)} \sim x^{-\gamma}\).
def measure_burstiness(inter_event_times):
"""
Calculate the burstiness index for a sequence of inter-event times.
The burstiness index is defined as:
B = (sigma - mu) / (sigma + mu)
where:
- sigma is the standard deviation of the inter-event times.
- mu is the mean of the inter-event times.
Parameters:
----------
inter_event_times : array-like
A sequence of inter-event times (time differences between consecutive events).
Returns:
-------
float
The burstiness index, where:
- B = -1 indicates perfectly regular events.
- B = 0 indicates Poissonian/random events.
- B > 0 indicates bursty events.
Raises:
------
ValueError
If the input array is empty or has insufficient data to calculate burstiness.
"""
# Ensure the input is a valid array
if len(inter_event_times) < 2:
raise ValueError("At least two inter-event times are required to calculate burstiness.")
# Compute mean and standard deviation
mu = np.mean(inter_event_times)
sigma = np.std(inter_event_times)
# Avoid division by zero
if sigma + mu == 0:
return 0.0
# Calculate and return the burstiness index
return (sigma - mu) / (sigma + mu)
print("burstiness index = %.6f" % measure_burstiness(ie_time_list))
burstiness index = 0.810840
ts_length = 100000
# random_time_list = np.random.randint(0,100,ts_length)
random_time_list = np.random.poisson(size=ts_length)
periodic_time_list = [10] * ts_length
powerlaw_time_list = np.random.zipf(2.0, ts_length)
print("Periodic inter-event times:\tBurstiness = %.3f"%measure_burstiness(periodic_time_list))
print("Random inter-event times:\tBurstiness = %.3f"%measure_burstiness(random_time_list))
print("Powerlaw inter-event times:\tBurstiness = %.3f"%measure_burstiness(powerlaw_time_list))
Periodic inter-event times: Burstiness = -1.000
Random inter-event times: Burstiness = 0.001
Powerlaw inter-event times: Burstiness = 0.980
Activity-Driven Network Model (if there’s time!)#
The Activity-Driven Network Model is a widely used framework for studying dynamic networks, where the structure evolves over time based on the intrinsic activity levels of nodes. Unlike static networks, where connections remain fixed, activity-driven networks capture the temporal nature of interactions, making them suitable for analyzing systems such as social interactions, communication patterns, and information diffusion.
Key Features of the Model#
The activity-driven network model assumes that each node in the network is characterized by an activity rate \( a_i \), which determines the probability that the node becomes active in a given time step. When a node becomes active, it forms temporary edges with other nodes in the network. These edges exist only for the duration of the time step, reflecting the transient nature of interactions in many real-world systems.
At each time step:
Node Activation: Each node becomes active with a probability proportional to its activity rate \( a_i \).
Edge Formation: Active nodes form \( m \) connections with other randomly selected nodes in the network.
Temporal Graph: The resulting network for the time step is recorded as a temporal graph.
Aggregation: Over multiple time steps, an aggregated network is constructed, representing all the edges formed throughout the simulation.
Model Parameters#
The activity-driven network model is controlled by the following parameters:
\( N \): The number of nodes in the network.
\( T \): The number of time steps over which the network evolves.
\( m \): The number of edges each active node creates per time step.
\( a_i \): The activity rate of node \( i \), sampled from a specified activity distribution (e.g., uniform or power-law).
The choice of the activity distribution is crucial, as it can reflect heterogeneous or homogeneous activity patterns. For example:
A uniform distribution implies that all nodes have similar probabilities of becoming active.
A power-law distribution assigns higher activity rates to a few nodes, reflecting real-world systems where a small subset of nodes drives most interactions.
Applications#
The activity-driven network model is versatile and has been applied in various domains:
Social Networks: Simulating interactions in human systems, such as email exchanges or face-to-face contact.
Epidemiology: Modeling the spread of infectious diseases, where temporary connections between individuals facilitate disease transmission.
Infrastructure: Understanding communication or transportation networks, where connections appear dynamically over time.
Information Diffusion: Capturing how information propagates through dynamic networks.
By explicitly modeling the temporal dynamics of networks, this approach provides a powerful tool for studying processes that unfold over time, such as epidemic outbreaks, cascades, and synchronization phenomena.
N nodes
each node has an activity potential \(x\), extracted from a distribution \(F(x)\)
Model dynamics:#
At each time step \(t\) the network \(G_t\) starts with \(N\) disconnected nodes
The node \(i\) fires with probability \(a_i \Delta t\), where \(a_i = \eta x_i\) and \(\eta\) is a recaling factor
Each active node generates \(m\) links connecting to \(m\) other randomly selected nodes (could be either active or non-active)
At next time step \(t+\Delta t\), all links in \(G_t\) are deleted. Go to step I.
Notes:
\(a_i = \eta x_i\) : Probability per unit time to create new connections with other nodes
\(\epsilon \leq x_i \leq 1 \)
The average number of active nodes per unit time is \(\eta \langle x \rangle N\)
Therefore:
Total number of edges per unit time: \(E_t = mN\eta\langle x \rangle\)
The average degree per unit time: $\(k_t = \frac{2E_t}{N} = 2m\eta\langle x \rangle\)$
At time \(T\), on the aggregated network up to time \(T\), the degree of node \(i\): $\(k_i(T)=N(1-e^{Tm\eta x_i/N})\)$
So the degree distribution for the aggregated network up to time \(T\): $\(P_T(k) \sim F\Big [ \frac{k}{Tm\eta} \Big ]\)$
References
Perra, N., Gonçalves, B., Pastor-Satorras, R. et al. Activity driven modeling of time varying networks. Sci Rep 2, 469 (2012). https://doi.org/10.1038/srep00469
Perra, N., Baronchelli, A., Mocanu, D., Gonçalves, B., Pastor-Satorras, R., & Vespignani, A. (2012). Random walks and search in time-varying networks. Physical Review Letters, 109(23), 238701. https://doi.org/10.1103/PhysRevLett.109.238701
def activity_driven_network(N, T, m, activity_distribution, plot=False):
"""
Simulate an activity-driven network model.
Parameters:
----------
N : int
Number of nodes in the network.
T : int
Number of time steps to simulate.
m : int
Number of edges each active node forms at each time step.
activity_distribution : callable
A function that generates activity rates for the nodes.
Example: lambda: np.random.uniform(0, 1) for uniform distribution.
plot : bool, optional
Whether to plot the final aggregated network. Default is False.
Returns:
-------
G_agg : networkx.Graph
The aggregated graph over all time steps.
G_t_list : list of networkx.Graph
A list of temporal graphs (one for each time step).
"""
# Step 1: Initialize nodes with activity rates
activity_rates = np.array([activity_distribution() for _ in range(N)])
nodes = list(range(N))
# Step 2: Create an empty aggregated graph
G_agg = nx.Graph()
# Step 3: Initialize list to store temporal graphs
G_t_list = []
# Step 4: Simulate the network over T time steps
for t in range(T):
# Create a new graph for the current time step
G_t = nx.Graph()
G_t.add_nodes_from(nodes)
# Determine active nodes
active_nodes = [node for node in nodes if np.random.rand() < activity_rates[node]]
# Create connections for each active node
for node in active_nodes:
# Choose m distinct nodes to connect to
targets = np.random.choice(nodes, size=m, replace=False)
for target in targets:
G_t.add_edge(node, target)
G_agg.add_edge(node, target) # Update the aggregated graph
# Store the temporal graph for the current time step
G_t_list.append(G_t)
return G_agg, G_t_list
# Example parameters
N = 100 # Number of nodes
T = 50 # Number of time steps
m = 3 # Number of edges per active node
activity_distribution = lambda: np.random.uniform(0.1, 0.5) # Uniform distribution of activity rates
# Simulate the activity-driven network
G_agg, G_t_list = activity_driven_network(N, T, m, activity_distribution, plot=True)
# Print basic stats
print(f"Aggregated Network: {G_agg.number_of_nodes()} nodes, {G_agg.number_of_edges()} edges")
print(f"Temporal Graphs: {len(G_t_list)} time steps")
Aggregated Network: 100 nodes, 3049 edges
Temporal Graphs: 50 time steps
Next time…#
Spatial Data, OSMNX, GeoPandas class_24_spatial.ipynb
References and further resources:#
Class Webpages
Jupyter Book: https://asmithh.github.io/network-science-data-book/intro.html
Syllabus and course details: https://brennanklein.com/phys7332-fall24
Cattuto, C., Van den Broeck, W., Barrat, A., Colizza, V., Pinton, J. F., & Vespignani, A. (2010). Dynamics of person-to-person interactions from distributed RFID sensor networks. PloS One, 5(7), e11596. https://doi.org/10.1371/journal.pone.0011596
Starnini, M., Baronchelli, A., & Pastor-Satorras, R. (2013). Modeling human dynamics of face-to-face interaction networks. Physical Review Letters, 110(16), 168701. https://doi.org/10.1103/PhysRevLett.110.168701
Barabási, AL. The origin of bursts and heavy tails in human dynamics. Nature 435, 207–211 (2005). https://doi.org/10.1038/nature03459
Perra, N., Gonçalves, B., Pastor-Satorras, R. et al. Activity driven modeling of time varying networks. Sci Rep 2, 469 (2012). https://doi.org/10.1038/srep00469
Perra, N., Baronchelli, A., Mocanu, D., Gonçalves, B., Pastor-Satorras, R., & Vespignani, A. (2012). Random walks and search in time-varying networks. Physical Review Letters, 109(23), 238701. https://doi.org/10.1103/PhysRevLett.109.238701
Barabási, AL. The origin of bursts and heavy tails in human dynamics. Nature 435, 207–211 (2005). https://doi.org/10.1038/nature03459
Goh, K. I., & Barabási, A. L. (2008). Burstiness and memory in complex systems. Europhysics Letters, 81(4), 48002. https://iopscience.iop.org/article/10.1209/0295-5075/81/48002
Karsai, M., Kaski, K., Barabási, AL. et al. Universal features of correlated bursty behaviour. Sci Rep 2, 397 (2012). https://doi.org/10.1038/srep00397